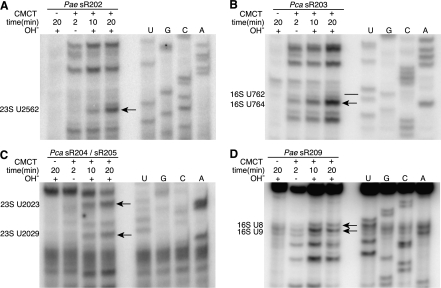
FIGURE 4.
Selected pseudouridine assays in Pyrobaculum aerophilum (Pae) and Pyrobaculum calidifontis (Pca) rRNA. In each experiment, four samples are treated with (+) or without (−) CMCT for either 2, 10, or 20 min followed with (+) or without (−) an alkaline (OH−) wash (lanes 1–4). RNA sequencing ladder is labeled in lanes U, G, C, A. The location of pseudouridine modification evidence is shown (black arrow), established by band absence in CMC− lane, faint banding at 2 min, and increasing band strength seen at 10- and 20-min intervals. Modification sites at 23S-U2582 (sR202), U2023 (sR204), and U2029 (sR205) show characteristic modification bands (panels A,C) located one position 3′ to the actual site of modification (lower in gel image) due to adduct formation. A potential site for sR203-guided modification (panel B) is U764 (black arrow), adjacent to the predicted U762 site that was negative for pseudouridine (black line). The modification site for sR209, 16S-U8 (panel D) is shown with the additional modification at U9.