The role for inosine-containing hyperedited dsRNA (IU-dsRNA) in cells is poorly understood. Previous studies have shown that IU-dsRNA binds specifically to various components of cytoplasmic stress granules, as well as to other proteins such as Tudor Staphylococcal Nuclease (Tudor-SN). Here the authors show that Tudor-SN localizes to cytoplasmic stress granules in HeLa cells undergoing arsenite-induced oxidative stress, or following transfection with long dsRNA (poly[IC]), which initiates an interferon cascade. They also demonstrate a novel interaction between Tudor-SN and ADAR1 and show that ADAR1 is also localized to stress granules in HeLa cells following various stresses.
Keywords: ADAR, Tudor-SN, stress granules, editing, dsRNA
Abstract
Hyperediting by adenosine deaminases that acts on RNA (ADARs) may result in numerous Adenosine-to-Inosine (A-to-I) substitutions within long dsRNA. However, while countless RNAs may undergo hyperediting, the role for inosine-containing hyperedited dsRNA (IU-dsRNA) in cells is poorly understood. We have previously shown that IU-dsRNA binds specifically to various components of cytoplasmic stress granules, as well as to other proteins such as Tudor Staphylococcal Nuclease (Tudor-SN). Tudor-SN has been implicated in diverse roles in mammalian cells, including transcription, splicing, RNAi, and degradation. Moreover, we have shown that Tudor-SN interacts directly with stress granule proteins. Here we show that Tudor-SN localizes to cytoplasmic stress granules in HeLa cells undergoing arsenite-induced oxidative stress, or following transfection with long dsRNA (poly[IC]), which initiates an interferon cascade. We additionally demonstrate a novel interaction between Tudor-SN and ADAR1. Finally, we show that ADAR1 is also localized to stress granules in HeLa cells following various stresses.
INTRODUCTION
Adenosine deaminases acting on RNA (ADARs) catalyze the deamination of adenosine (A) to inosine (I) within double-stranded RNA (dsRNA) (Bass 2002; Valente et al. 2005). Of the three ADARs (ADAR1–3) described in mammals, only ADAR1 and ADAR2 appear to be catalytically active (Bass 2002; Valente et al. 2005). Two major isoforms of ADAR1 exist in mammalian cells; ADAR1 p110 is constitutively expressed and is predominantly nuclear, while ADAR1 p150 is inducible by interferon (George and Samuel 1999) and shuttles between the nucleus and cytoplasm (Eckmann et al. 2001). Analysis of ADAR1- and ADAR2-null mutants has underlined their functional importance (Higuchi et al. 2000; Wang et al. 2004; Hartner et al. 2009).
A-to-I editing may occur selectively within mRNA, which can result in codon changes as inosine is interpreted as guanosine by the translation machinery. ADARs may additionally hyperedit long dsRNAs, which can result in up to 50% of A residues being changed to I. Moreover, hyperedited inosine-containing dsRNAs (IU-dsRNA) are likely to have localized changes in their RNA structure due to the relative instability of IU pairs (Serra et al. 2004). Most mammalian editing occurs within noncoding regions of RNA, particularly within Alu repeat sequences (Morse et al. 2002; Blow et al. 2004; Levanon et al. 2004; Barak et al. 2009). However, while numerous RNAs may be extensively edited, the possible functions of IU-dsRNA in cells are not fully understood. Various studies have suggested diverse fates for IU-dsRNA (Scadden and Smith 2001; Scadden 2005, 2007; Chen et al. 2008; Hundley et al. 2008; Chen and Carmichael 2009).
We have previously shown that IU-dsRNA specifically binds a protein complex that largely comprises components of cytoplasmic stress granules (Scadden 2007; Anderson and Kedersha 2009). In addition, we have shown that IU-dsRNA interacts specifically with Tudor Staphylococcal Nuclease (Tudor-SN) (Scadden 2005). Tudor-SN appears to have diverse functions in mammalian cells. It was first identified as a transcriptional coactivator of EBNA-2 (Callebaut and Mornon 1997), and subsequently of STAT5 and STAT6 (Yang et al. 2002; Paukku et al. 2003). Tudor-SN was later shown to interact with snRNP proteins and facilitates spliceosome assembly (Yang et al. 2007). Tudor-SN also appears to have cytoplasmic roles, in that it has been described a component of RISC (Caudy et al. 2003), and enhances cleavage of IU-dsRNA (Scadden 2005). We have additionally shown that Tudor-SN interacts specifically with the stress granule proteins that bind IU-dsRNA (Scadden 2007).
Formation of stress granules provides eukaryotic cells with a means of survival during stress (Anderson and Kedersha 2008, 2009). Stress granules typically form as a result of translation inhibition, and are therefore comprised of stalled initiation complexes and small ribosomal proteins (Kedersha et al. 2002). Importantly, sequestration of a set of translationally arrested mRNAs allows selective synthesis of proteins required for cell survival (Anderson and Kedersha 2008, 2009). When stress conditions are removed, sequestered mRNAs may reassemble on polysomes and resume translation (Anderson and Kedersha 2008). Alternatively, mRNAs may be degraded within cytoplasmic processing bodies (P-bodies), which are dynamically linked to stress granules (Kedersha et al. 2005).
Here we present experimental data demonstrating that Tudor-SN specifically localizes to stress granules in HeLa cells subject to either arsenite-induced oxidative stress or following transfection of long dsRNA (poly[IC]), which mimics viral infection. Moreover, we showed that a pair of staphylococcal nuclease domains was sufficient for localization. In contrast, Tudor-SN was not detected in P-bodies. Our data additionally revealed a novel interaction between Tudor-SN and ADAR1. In light of this observation, we showed that ADAR1 was also localized to stress granules in HeLa cells following either oxidative stress or transfection of poly(IC).
RESULTS
Previously we have shown that IU-dsRNA specifically binds a protein complex that largely comprised components of cytoplasmic stress granules (Scadden 2007). However, other proteins that had not previously been associated with stress granules also interacted specifically with IU-dsRNA. One such protein was Tudor-SN, which appears to play a variety of roles in mammalian cells (Callebaut and Mornon 1997; Yang et al. 2002; Scadden 2005; Yang et al. 2007). We therefore set out to determine whether Tudor-SN was also associated with stress granules during various cellular stresses.
Tudor-SN interacts with stress granule proteins
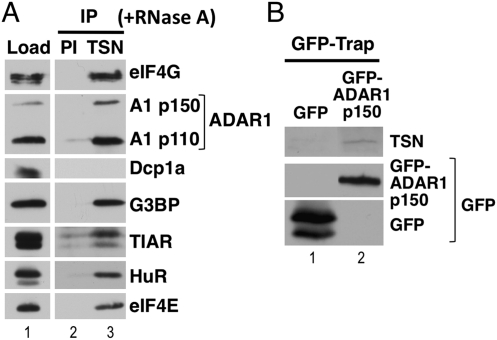
We initially carried out experiments to confirm that Tudor-SN interacted specifically with various stress granule proteins. A Tudor-SN antibody (TSN) was therefore used to immunoprecipitate proteins from RNase A-treated HeLa cell lysate. Pre-immune (PI) serum was used as a control. The immunoprecipitates were subsequently analyzed by immunoblotting (Fig. 1A). These data confirmed that Tudor-SN interacted specifically with a variety of stress granule proteins (eIF4G, G3BP, TIAR, HuR, and eIF4E), in an RNA-independent manner (Fig. 1A, cf. lanes 2 and 3). In contrast, Tudor-SN did not interact with Dcp1, which is a component of cytoplasmic P-bodies rather than stress granules (Anderson and Kedersha 2009). Due to the previously described interaction between Tudor-SN and IU-dsRNA (Scadden 2005), we speculated that Tudor-SN may also interact with ADARs. The immunoblot was therefore probed with an antibody specific for ADAR1 (ADAR1), which confirmed that ADAR1 and Tudor-SN specifically interacted in HeLa cell lysates (Fig. 1A). We subsequently verified the interaction using reciprocal immunoprecipitation experiments. HeLa cells were transfected with an expression construct for GFP–ADAR1 p150 or GFP (control), and lysates were prepared after 24 h. GFP–ADAR1 p150 or GFP were immunoprecipitated using GFP–Trap, and the immunoprecipitates subsequently analyzed by immunoblotting (Fig. 1B). A GFP antibody was initially used to verify expression of GFP and GFP–ADAR1 p150. The immunoblot was subsequently probed with a specific Tudor-SN antibody (TSN), which confirmed that Tudor-SN interacted specifically with GFP–ADAR1 p150 (Fig. 1B, cf. lanes 1 and 2).
FIGURE 1.
Tudor-SN interacts with stress granule components. (A) A Tudor-SN (TSN) antibody was used to immunoprecipitate proteins from RNase A-treated HeLa cell lysates, and immunoblotting was subsequently used to detect interacting proteins. Pre-immune serum (PI) was used as a control. Total protein (Load) is shown. (B) HeLa cells were transiently transfected with expression vectors for GFP (control) or GFP–ADAR1 p150, and lysates prepared after 24 h. GFP–Trap was subsequently used to immunoprecipitate GFP or GFP–ADAR1 p150, and immunoblotting used to analyze the immunoprecipitates. A specific Tudor-SN antibody (TSN) was used to detect Tudor-SN. The immunoblot was additionally probed with anti-GFP to verify expression of GFP and GFP–ADAR1 p150.
Our data conclusively demonstrated that Tudor-SN was found in a complex with various stress granule proteins, as well as showing a novel interaction with ADAR1.
Tudor-SN localizes to stress granules in arsenite-treated HeLa cells
We have demonstrated a specific interaction between Tudor-SN and stress granule proteins. In light of these findings, we next asked whether Tudor-SN was localized to stress granules in HeLa cells subject to arsenite-induced oxidative stress.
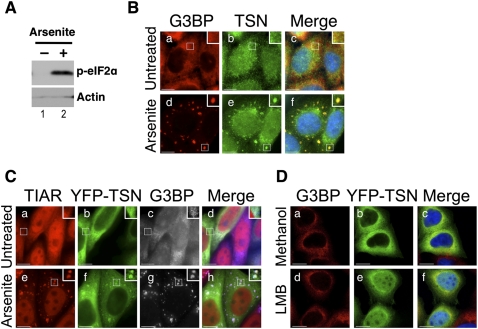
HeLa cells were grown in the absence or presence of 0.5 mM arsenite for 30 min, and then allowed to recover for an additional 30 min in the absence of arsenite. These conditions for arsenite treatment were also used in subsequent experiments. Immunoblotting was used to verify that arsenite treatment resulted in phosphorylation of eIF2α (Fig. 2A, p-eIF2α), which typically precedes stress granule formation. Cells were subsequently fixed and stained for endogenous Tudor-SN and G3BP, a stress granule marker, using specific antibodies (Fig. 2B). Secondary antibodies were used to detect Tudor-SN (green) and G3BP (red), which were visualized using fluorescence microscopy. The nucleus was stained with DAPI (blue). In the absence of arsenite (Untreated), localization of G3BP was cytoplasmic while Tudor-SN was distributed throughout the cell (Fig. 2B, a and b, respectively). In arsenite-treated cells, G3BP localized to cytoplasmic stress granules, as expected (Fig. 2B, d). Similarly, Tudor-SN also localized to cytoplasmic foci following arsenite treatment (Fig. 2B, e). Moreover, the merged image confirmed that Tudor-SN and G3BP colocalized to stress granules in arsenite-treated cells (Fig. 2B, f). This observation was verified using other stress granule markers (e.g., TIAR, HuR [data not shown]). These data therefore confirmed that endogenous Tudor-SN localized to stress granules in HeLa cells subject to arsenite-induced oxidative stress.
FIGURE 2.
Tudor-SN localizes to stress granules. (A) Lysates were prepared from HeLa cells treated ±arsenite, and immunoblotting was used to analyze phosphorylation of eIF2α (p-eIF2α). Actin was a loading control. (B) HeLa cells were cultured in the absence (a–c) or presence of arsenite (d–f), then fixed and processed for visualization of G3BP (red; a,d) and Tudor-SN (TSN) (green; b,e) using fluorescence microscopy. DAPI staining is blue (Merge; c,f). (C) HeLa cells were transiently transfected with an expression vector for YFP–TSN. After 24 h the cells were cultured in the absence (a–d) or presence of arsenite (e–h) before processing for visualization of TIAR (red; a,e), YFP–TSN (green; b,f), and G3BP (blue [shown as gray]; c,g) using fluorescence microscopy. Merged images are shown for untreated and arsenite-treated cells (Merge; d,h). (D) HeLa cells were transiently transfected with an expression vector for YFP–TSN. After 24 h the cells were cultured in the absence (Methanol; a–c) or presence of leptomycin B (LMB; d–f) before processing for visualization of G3BP (red; a,d) and YFP–TSN (green; b,e) using confocal microscopy. DAPI staining is blue (Merge; c,f). Bar, 10 μm.
We next tested whether full-length Tudor-SN fused to YFP (YFP–TSN; Fig. 4A, see below) was similarly localized to stress granules following arsenite treatment. HeLa cells were transfected with the YFP–TSN expression construct, and after 24 h were cultured with or without arsenite. Cells were subsequently fixed and processed for visualization of YFP–TSN (green), TIAR (red), and G3BP (blue; shown as gray for clarity) using fluorescence microscopy (Fig. 2C). In the absence of arsenite, YFP–TSN was predominantly cytoplasmic (Fig. 2C, b). While the localization of Tudor-SN appeared slightly different from the endogenous protein, this reflects the relatively high expression of recombinant Tudor-SN, and is consistent with previous observations (Gao et al. 2010). Nevertheless, when HeLa cells were treated with arsenite, TIAR, G3BP, and YFP–TSN colocalized to cytoplasmic stress granules (Fig. 2C, e–h). These data therefore demonstrate that recombinant Tudor-SN also localized to stress granules during oxidative stress.
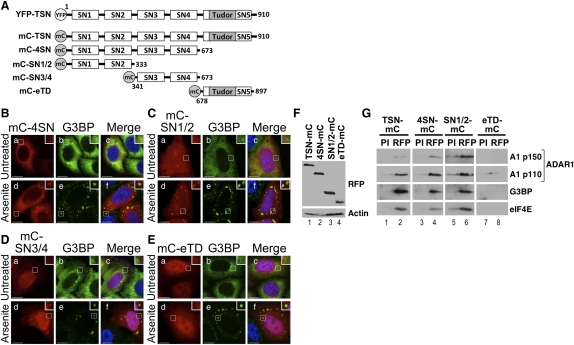
FIGURE 4.
Staphylococcal domains of Tudor-SN localize to stress granules. (A) A schematic diagram of the YFP–TSN and mC–TSN constructs; the amino acids of Tudor-SN included in each construct are shown. (B–E) HeLa cells were transiently transfected with expression vectors for (B) mC–4SN, (C) mC–SN1/2, (D) mC–SN3/4, and (E) mC–eTD. After 24 h the cells were cultured in the absence (a–c) or presence of arsenite (d–f) before processing for visualization of mC–4SN, mC–SN1/2, mC–SN3/4, or mC–eTD, respectively (red; a,d), and G3BP (green; b,e) using confocal microscopy. DAPI staining is in blue (Merge; c,f). Bar, 10 μm. (F) HeLa cells were transiently transfected with expression vectors for TSN–mC, 4SN–mC, SN1/2, and mC–eTD, and lysates prepared after 24 h. Immunoblotting was used to analyze expression. Actin was a loading control. (G) An RFP antibody was used to immunoprecipitate GFP–TSN proteins from HeLa cell lysates, and immunoblotting was subsequently used to detect interacting proteins. Pre-immune serum (PI) was used as a control.
Tudor-SN is a shuttling protein
Previous data showed that Tudor-SN was predominantly found in the cytoplasmic fraction of mammalian cells (Caudy et al. 2003). This is consistent with our findings that Tudor-SN localizes to cytoplasmic foci during stress. However, its nuclear roles in transcription and splicing (Callebaut and Mornon 1997; Yang et al. 2002; Paukku et al. 2003; Yang et al. 2007) suggest that Tudor-SN must also localize to the nucleus. Moreover, this idea is in agreement with our observation that endogenous Tudor-SN localizes to both the nucleus and cytoplasm in unstressed cells (e.g., Fig. 2B). We therefore investigated whether Tudor-SN shuttles between the nuclear and cytoplasmic compartments in HeLa cells.
HeLa cells were transiently transfected with the YFP–TSN expression construct, and after 24 h were cultured in the presence of leptomycin B (LMB) (Fig. 2D, d–f). LMB specifically inhibits nuclear export in human cells via direct binding to CRM1 (Kudo et al. 1998). HeLa cells were alternatively treated with methanol as a control (Fig. 2D, a–c). Cells were subsequently fixed and processed for visualization of YFP–TSN (green) and G3BP (red), which is primarily a cytoplasmic protein (Tourriere et al. 2001). DAPI (blue) was used to stain the nucleus. In methanol-treated (control) cells, G3BP and YFP–TSN colocalized in the cytoplasm (Fig. 2D, a–c). When HeLa cells were treated with LMB, G3BP remained cytoplasmic, as expected (Fig. 2D, d). In contrast, Tudor-SN was detected in both the nucleus and cytoplasm (Fig. 2D, e,f). These data therefore confirmed that Tudor-SN shuttles between the nucleus and cytoplasm in HeLa cells, which is presumably necessary in order to fulfill both nuclear and cytoplasmic functions.
Tudor-SN does not localize to P-bodies
While we have conclusively demonstrated that Tudor-SN colocalized with the stress granule proteins G3BP and TIAR following oxidative stress, we wanted to determine whether Tudor-SN was also found in cytoplasmic P-bodies (Anderson and Kedersha 2009).
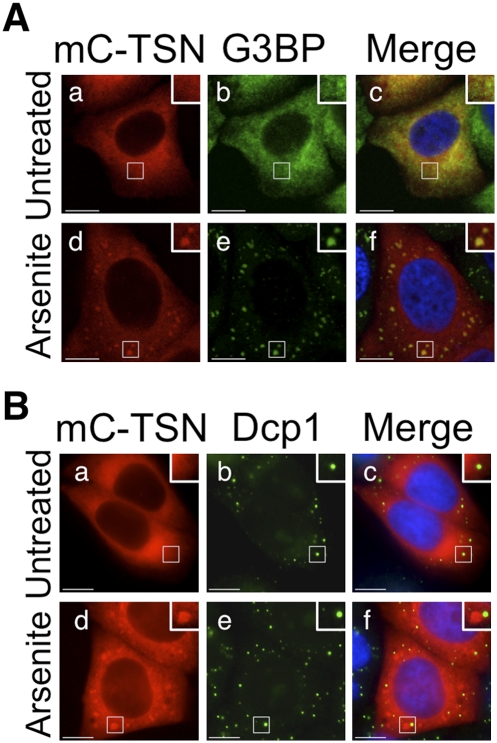
HeLa cells were transiently transfected with an expression construct encoding full-length Tudor-SN fused to mCherry (mC–TSN; Fig. 4A, see below). After 24 h cells were incubated with or without arsenite, then fixed and processed for visualization of various proteins using fluorescence microscopy. Localization of mCherry–Tudor-SN (mC–TSN; red) and G3BP (green) were initially analyzed using confocal microscopy (Fig. 3A). The nucleus was stained with DAPI (blue). In the absence of arsenite, both mC–TSN and G3BP were predominantly cytoplasmic (Fig. 3A, a–c). When HeLa cells were treated with arsenite, both G3BP and mC–TSN colocalized to stress granules (Fig. 3A, d–f). HeLa cells expressing mC–TSN were additionally stained with antibodies against Dcp1a, which localizes to P-bodies (Fig. 3B, b,e [green]). In arsenite-treated cells, Tudor-SN and Dcp1a localized to distinct cytoplasmic foci corresponding to stress granules and P-bodies, respectively (Fig. 3B, d–f). Moreover, in keeping with previous studies, the stress granules observed were frequently juxtaposed with P-bodies (Fig. 3B, f; Kedersha et al. 2005). These data therefore confirmed that Tudor-SN localized to stress granules but not P-bodies during oxidative stress.
FIGURE 3.
Tudor-SN does not localize to P-bodies. (A) HeLa cells were transiently transfected with an expression vector for mC–TSN. After 24 h the cells were cultured in the absence (a–c) or presence of arsenite (d–f) before processing for visualization of mC–TSN (red; a,d) and G3BP (green; b,e) using confocal microscopy. DAPI staining is blue (Merge; c,f). (B) HeLa cells were transiently transfected with an expression vector for mC–TSN. After 24 h the cells were cultured in the absence (a–c) or presence of arsenite (d–f) before processing for visualization of mC–TSN (red; a,d) and Dcp-1 (green; b,e) using fluorescence microscopy. DAPI staining is blue (Merge; c,f). Bar, 10 μm.
Staphylococcal nuclease domains localize Tudor-SN to stress granules
We next investigated which domains of Tudor-SN were responsible for localization to stress granules. For this analysis, expression constructs were prepared where mCherry was fused to various domains of Tudor-SN, according to the predicted domain boundaries (ExPASy PROSITE; Sigrist et al. 2010; Fig. 4A). HeLa cells were transfected with the various mCherry–TSN expression constructs (mC–4SN, mC–SN1/2, mC–SN3/4, mC–eTD), and after 24 h were cultured with or without arsenite. Immunoblotting using a mCherry antibody confirmed that expression was similar for each construct (data not shown). Cells were subsequently fixed and processed for visualization of the mCherry–Tudor-SN domains (red) and G3BP (green) using confocal microscopy (Fig. 4B–E). The nucleus was stained with DAPI (blue). In the absence of arsenite, mC–4SN colocalized with G3BP in the cytoplasm (Fig. 4B, a–c). In contrast, mC–SN1/2, mC–SN3/4, and mC–eTD were predominantly nuclear in untreated cells (Fig. 4C–E, a). Following treatment of HeLa cells with arsenite, mC–4SN, mC–SN1/2, and mC–SN3/4 colocalized with G3BP in stress granules (Fig. 4B,C, d–f). In contrast, mC–eTD was not detected in stress granules following arsenite treatment (Fig. 4E, d–f). Equivalent observations were made using Flag-tagged Tudor-SN domains (data not shown). These data together suggested that staphylococcal nuclease domains 1–4 were responsible for localization of Tudor-SN to stress granules. Furthermore, we have demonstrated that the presence of only two staphylococcal nuclease domains (SN1/2 or SN3/4) is sufficient for localization to stress granules.
We next tested which of the Tudor-SN domains interacted with different stress granule proteins and ADAR1. HeLa cells were transfected with various TSN–mCherry expression constructs (TSN–mC, 4SN–mC, SN1/2–mC, eTD–mC) and lysates prepared after 24 h. Immunoblotting using an RFP antibody confirmed that expression was similar for each construct (Fig. 4F). Actin was a loading control. The RFP antibody was subsequently used to immunoprecipitate the various TSN–mC domains, where PI serum was used as a control. Immunoblots were subsequently used to analyze the interacting proteins (Fig. 4G). These data revealed that the TSN–mC, 4SN–mC, and SN1/2–mC immunoprecipitates were greatly enriched for ADAR1, G3BP, and eIF4E, relative to the control (Fig. 4G, cf. lanes 1 and 2, 3 and 4, 5 and 6). In contrast, the eTD–mC domain did not immunoprecipitate these proteins (Fig. 4G, lanes 7,8). These data together suggested that the first four SN domains of Tudor-SN were important for interacting with various stress granule proteins. Moreover, these findings were consistent with the observation that staphylococcal nuclease domains 1–4 were responsible for localization of Tudor-SN to stress granules (Fig. 4B–E).
Tudor-SN localizes to poly(IC)-induced stress granules
The composition of stress granules that assemble in response to different stresses can vary (Kedersha et al. 2005; Buchan and Parker 2009; Buchan et al. 2011). We have demonstrated that Tudor-SN localizes to stress granules induced by oxidative stress. We next tested whether Tudor-SN also localized to stress granules induced by long dsRNA (poly[IC]), which mimics viral infection. Viral infection typically induces stress granules via activation of PKR (protein kinase R), a double-stranded RNA-dependent kinase, which phosphorylates eIF2α and thereby inhibits translation (Anderson and Kedersha 2002).
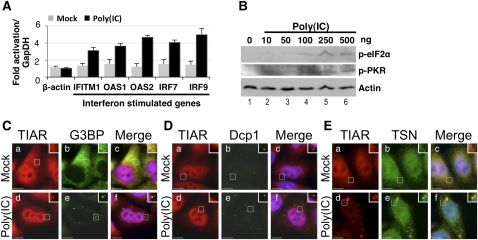
HeLa cells were mock-transfected or transfected with poly(IC). We initially showed that transfection of HeLa cells was efficient by analyzing expression of interferon stimulated genes (ISGs), which are up-regulated by poly(IC) (Stark et al. 1998). RNA was thus harvested after 7 h, and RT/qPCR was used to quantify expression of ISGs (IFITM1, OAS2, OAS1, IRF7, IRF9) or a control gene (β-actin) (Fig. 5A). Fold-change in mRNA levels were calculated relative to those in mock-transfected cells, and normalized to GapDH. This analysis confirmed that expression of all ISGs tested was increased in the presence of poly(IC) but not in mock-transfected cells (Fig. 5A), thereby verifying that transfection of cells by poly(IC) was efficient. This observation was supported by immunostaining poly(IC)-transfected cells with an antibody specific for dsRNA, which confirmed that 64.1 ± 5.7% (n = 6) of HeLa cells contained poly(IC) (data not shown).
FIGURE 5.
Stress granules are induced by poly(IC). (A) HeLa cells were mock-transfected or transfected with poly(IC), and RT/qPCR was used to quantify expression of β-actin or ISGs (IFITM1, OAS2, OAS1, IRF7, and IRF9) after 7 h. Fold-change in mRNA levels were calculated relative to those without poly(IC) (mock), and normalized to GapDH. Error bars are mean ± s.d. (n ≥ 3). (B) HeLa cells were transfected with 0–500 ng poly(IC), and cell lysates were prepared after 7 h. Immunoblotting was used to analyze phosphorylation of eIF2α (p-eIF2α) and PKR (p-PKR). Actin was a loading control. (C–E) HeLa cells were transfected with poly(IC), then fixed and processed after 7 h cells for immunofluorescence microscopy. DAPI staining is blue (Merge; c,f). (C) HeLa cells were stained for TIAR (red; a,d) and G3BP (green; b,e). (D) HeLa cells were stained for TIAR (red; a,d) and Dcp1 (green; b,e). (E) HeLa cells were stained for TIAR (red; a,d) and Tudor-SN (TSN) (green; b,e). Bar, 10 μm.
We additionally verified that eIF2α and PKR were phosphorylated in the presence of poly(IC). HeLa cells were transfected with 0–500 ng poly(IC), and lysates prepared after 7 h. Immunoblots were subsequently used to demonstrate that both eIF2α (p-eIF2α) and PKR (p-PKR) underwent phosphorylation, hence activation, with increasing concentrations of poly(IC) (Fig. 5B). Actin was a loading control.
Finally we analyzed stress granules using immunofluorescence. HeLa cells were mock-transfected or transfected with poly(IC), and after 7 h cells were fixed and processed for visualization of various proteins using fluorescence microscopy. Cells were initially stained for stress granule markers TIAR (red) and G3BP (green) to confirm that stress granules were induced by poly(IC) (Fig. 5C). DAPI (blue) was used to stain the nucleus. In mock-transfected cells, TIAR was distributed throughout the cell while G3BP localized to the cytoplasm (Fig. 5C, a–c). Following transfection of HeLa cells with poly(IC), both TIAR and G3BP colocalized in discrete cytoplasmic foci (Fig. 5C, d–f). Cells were additionally stained with antibodies for Dcp1a (green) or TIAR (red) (Fig. 5D). These data confirmed that the poly(IC)-induced TIAR-containing foci were distinct from the Dcp1a-containing P-bodies (Fig. 5D, d–f). These data were therefore consistent with the formation of cytoplasmic stress granules in response to poly(IC). Finally, cells were stained with antibodies against TIAR (red) and Tudor-SN (green) in order to determine whether Tudor-SN also localized to poly(IC)-induced stress granules (Fig. 5E). In the absence of poly(IC), both TIAR and Tudor-SN were distributed throughout the cell (Fig. 5E, a–c). Following transfection with poly(IC), both TIAR and Tudor-SN colocalized to cytoplasmic stress granules (Fig. 5E, d–f). These data therefore confirmed that Tudor-SN additionally localized to stress granules that assembled in response to long dsRNA.
ADAR1 localizes to stress granules
We have shown that Tudor-SN interacts specifically with ADAR1 (Fig. 1), and that Tudor-SN localizes to stress granules (Figs. 2–5). We therefore investigated whether ADAR1 additionally localizes to stress granules in HeLa cells following arsenite treatment or transfection of long dsRNA. As mentioned above, two major isoforms of ADAR1 (p110 and p150) exist in mammalian cells. Previous data have shown that, although the interferon-inducible ADAR1 p150 shuttles between the nucleus and cytoplasm, it is largely cytoplasmic. In contrast, ADAR1 p110 is predominantly nuclear, although it can localize to the cytoplasm in the absence of transcription (Eckmann et al. 2001; Strehblow et al. 2002; Desterro et al. 2003). In view of its cellular localization, we have focused on the cytoplasmic isoform of ADAR1 (p150) in our analyses.
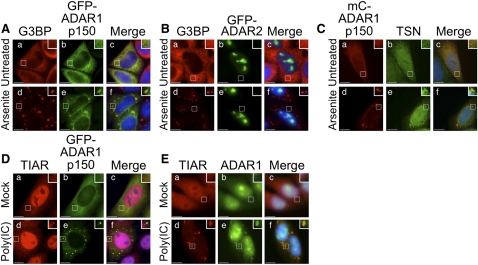
We first tested whether GFP–ADAR1 p150 localized to stress granules during oxidative stress. HeLa cells were transiently transfected with plasmids expressing either GFP–ADAR1 p150 or GFP–ADAR2. GFP–ADAR2 was used as a control as its expression was exclusively nuclear (Desterro et al. 2003). After 24 h cells were cultured in the presence or absence of arsenite, then fixed and processed for visualization of GFP–ADAR1 p150 or GFP–ADAR2 (green), or G3BP (red) using fluorescence microscopy (Fig. 6A,B). In the absence of arsenite treatment, GFP–ADAR1 p150 was predominantly cytoplasmic (Fig. 6A, b) while GFP–ADAR2 localized to the nucleoli, as reported previously (Fig. 6B, b; Desterro et al. 2003). Localization of G3BP was cytoplasmic (Fig. 6A,B, a). When HeLa cells were treated with arsenite, GFP–ADAR1 p150 colocalized with G3BP to cytoplasmic stress granules (Fig. 6A, d–f). In contrast, GFP–ADAR2 remained in the nucleus following arsenite treatment (Fig. 6B, d–f). These data together therefore confirmed that ADAR1 p150 localized to stress granules that resulted from oxidative stress.
FIGURE 6.
ADAR1 localizes to stress granules. (A) HeLa cells were transiently transfected with an expression vector for GFP–ADAR1 p150. After 24 h cells were cultured in the absence (a–c) or presence of arsenite (d–f) before processing for visualization of G3BP (red; a,d) or GFP–ADAR1 p150 (green; b,e). DAPI staining is blue (Merge; c,f). (B) HeLa cells were transiently transfected with an expression vector for GFP–ADAR2. After 24 h the cells were cultured in the absence (a–c) or presence of arsenite (d–f) before processing for visualization of G3BP (red; a,d) and GFP–ADAR2 (green; b,e). DAPI staining is blue (Merge; c,f). (C) HeLa cells were transiently transfected with an expression vector for mC–ADAR1 p150. After 24 h the cells were cultured in the absence (a–c) or presence of arsenite (d–f) before processing for visualization of mC–ADAR1 p150 (red; a,d) and TSN (green; b,e). DAPI staining is blue (Merge; c,f). (D) HeLa cells were transiently transfected with an expression vector for GFP–ADAR1 p150. After 24 h the cells were transfected with (d–f) or without (a–c) poly(IC). After 7 h, cells were fixed and immunofluorescence microscopy was used to visualize TIAR (red; a,d) and GFP–ADAR1 p150 (green; b,e). DAPI staining is blue (Merge; c,f). (E) HeLa cells were transfected with (d–f) or without (a–c) poly(IC). After 7 h, cells were fixed and immunofluorescence microscopy was used to visualize TIAR (red; a,d) and endogenous ADAR1 (green; b,e). DAPI staining is blue (Merge; c,f). Bar, 10 μm.
We additionally analyzed whether ADAR1 p150 and Tudor-SN colocalized during oxidative stress. HeLa cells were transfected with an expression construct for mCherry–ADAR1 p150 (mC–ADAR1 p150), and after 24 h were cultured in the presence or absence of arsenite. Cells were subsequently fixed and processed for visualization of mC–ADAR1 p150 (red) or endogenous Tudor-SN (TSN; green) using fluorescence microscopy (Fig. 6C). In the absence of arsenite treatment, mC–ADAR1 p150 was largely cytoplasmic (Fig. 6C, a) while Tudor-SN was distributed throughout the cell (Fig. 6C, b). When HeLa cells were treated with arsenite, mC–ADAR1 p150 colocalized with Tudor-SN in cytoplasmic stress granules (Fig. 6C, d–f).
We next tested whether ADAR1 p150 localized to stress granules induced by transfection of long dsRNA (poly[IC]). HeLa cells were transfected with the expression construct for GFP–ADAR1 p150, and after 24 h were mock-transfected or transfected with poly(IC). After a further 7 h cells were fixed for visualization of TIAR (red) or GFP–ADAR1 p150 (green) using fluorescence microscopy (Fig. 6D). In the absence of poly(IC) (Mock), TIAR was mainly nuclear (Fig. 6D, a) while GFP–ADAR1 p150 was predominantly cytoplasmic (Fig. 6D, b). In the presence of poly(IC), TIAR and GFP–ADAR1 p150 colocalized in the cytoplasm to stress granules (Fig. 6D, d–f). These data therefore confirmed that ADAR1 p150 localized to stress granules that assemble in response to long dsRNA.
Finally we tested whether endogenous ADAR1 also localized to stress granules that assembled following transfection of long dsRNA. HeLa cells were mock-transfected or transfected with poly(IC), then fixed after 7 h for visualization of TIAR (red) or ADAR1 (green) using fluorescence microscopy (Fig. 6E). As the ADAR1 antibody recognizes both ADAR1 p110 and ADAR1 p150 (Fig. 1A), both isoforms were detected in this analysis. In the absence of poly(IC), ADAR1 was distributed throughout the cell, although it was greatly enriched in the nucleus (Fig. 6E, b). Following transfection of poly(IC), TIAR localized to stress granules, as expected (Fig. 6C, d,f). Importantly, endogenous ADAR1 colocalized with TIAR in cytoplasmic stress granules following transfection of poly(IC) (Fig. 6E, e,f). Although these data did not allow us to determine whether ADAR1 p110 or p150 localized to stress granules, we speculate that this is most likely to be the interferon-inducible cytoplasmic ADAR1 p150 isoform. Moreover, this idea is in keeping with the observation that GFP–ADAR1 p150 localized to stress granules induced by poly(IC) (Fig. 6D). Regardless, these data confirmed that ADAR1 localized to stress granules that assemble in response to poly(IC).
Our data together allowed us to conclude that ADAR1 p150 localized to stress granules that formed in response to either arsenite-induced oxidative stress or transfection of long dsRNA.
DISCUSSION
In light of our previous findings that showed that Tudor-SN interacted directly with stress granule proteins (Scadden 2007), we investigated whether Tudor-SN localized to stress granules following cellular stress. Direct visualization of either endogenous or recombinant Tudor-SN using fluorescent microscopy confirmed that Tudor-SN was localized to stress granules that formed in HeLa cells due to either oxidative stress or interferon induction (Figs. 2–5). Moreover, these data were in agreement with other recent findings that also demonstrate that Tudor-SN localizes to stress granules during various stress conditions (Gao et al. 2010).
In the absence of cellular stress, localization of endogenous Tudor-SN throughout the cell (Fig. 2B) is consistent with its having both nuclear and cytoplasmic functions (Callebaut and Mornon 1997; Yang et al. 2002; Caudy et al. 2003; Paukku et al. 2003; Scadden 2005; Yang et al. 2007). Furthermore, by blocking nuclear export with LMB, we have shown that Tudor-SN shuttles between compartments (Fig. 2D). While much remains to be understood about the role of Tudor-SN in stress granules, Tudor-SN appears to be an integral component of stress granules that assemble following oxidative stress, heat-shock, or interferon induction (Figs. 1–5; Gao et al. 2010). In contrast, Tudor-SN was not found in P-bodies under any of the conditions tested (Figs. 3, 5). Insights into the function of Tudor-SN in stress granules may be gained by more precisely delineating the interactions between Tudor-SN and other stress granule proteins (Fig. 4). Defining the specific contacts made between Tudor-SN and other proteins may provide clues as to its exact function during stress.
Intriguingly, we have previously shown that both Tudor-SN and stress granule proteins interact specifically with IU-dsRNA (Scadden 2005, 2007). In view of these observations, we undertook experiments that revealed a novel interaction between ADAR1 and Tudor-SN (Fig. 1). Importantly, we demonstrated that the cytoplasmic GFP–ADAR1 p150 also localized to stress granules following either arsenite-induced oxidative stress or transfection of poly(IC) (Fig. 6). This significant finding supports previous data that suggests ADAR1 p150 plays an important role during various cellular stresses. While our data do not exclude the possibility that ADAR1 p110 may also localize to stress granules, this seems less likely in view of its predominantly nuclear role in mammalian cells (Bass 2002; Valente et al. 2005). However, other nuclear proteins relocalize to stress granules during stress conditions (e.g., HuR, TIAR) (Anderson and Kedersha 2008).
Expression of ADAR1 p150 in wild-type cells is induced by interferon, and in response to serum deprivation (George and Samuel 1999; Wang et al. 2004). These observations suggested that the ADAR1 p150 isoform is necessary for cell survival. In keeping with this idea, widespread apoptosis and defective haematopoiesis were observed in Adar1−/− mice, which resulted in early embryonic death (∼E11.5) (Wang et al. 2004; Hartner et al. 2009). Expression of ISGs was also up-regulated in ADAR1-null mice (Hartner et al. 2009). Increased apoptosis was additionally observed in fibroblasts derived from Adar1−/− mouse embryos following serum starvation (Wang et al. 2004). Viral infection of ADAR1-deficient HeLa cells also resulted in enhancing apoptosis (Toth et al. 2009). Our recent data showed that IU-dsRNA per se suppressed both apoptosis and induction of ISGs (Vitali and Scadden 2010). These data together reinforce the idea that ADAR1 p150 is crucial for suppression of apoptosis, hence cell survival.
Localization of ADAR1 p150 to stress granules following either arsenite-induced oxidative stress or treatment with poly(IC) is likely to be important for cell survival. It is tempting to speculate that ADAR1 edits a subset of RNAs sequestered within stress granules, and thereby generates IU-dsRNA that will subsequently suppress interferon induction and apoptosis. Cell survival will therefore be enhanced following recruitment of ADAR1 to stress granules. ADAR1 p150 may alternatively edit target RNAs elsewhere in the cytoplasm, and the resultant IU-dsRNA may trigger stress granule formation following interaction with the stress-granule-like complex (Scadden 2007). In this instance, ADAR1 may localize to stress granules by virtue of association with the edited RNA. Regardless, the observation that ADAR1 localizes to stress granules is consistent with the idea that ADAR1 is involved in various stress responses, as outlined above.
We now intend to undertake experiments that will more precisely define the role of ADAR1 in stress granules. While some proteins appear within stress granules by “piggy-back” association with core stress granule components, other proteins nucleate stress granule assembly or serve as molecular scaffolds that define the SG domain (e.g., TIAR, G3BP) (Anderson and Kedersha 2008). It will now be important to determine what role ADAR1 p150 plays in stress granules, and how its presence in stress granules may provide a link with other stress response pathways. Characterizing the function of ADAR1 in stress granules will help to elucidate the crucial role of ADAR1 in cellular stress responses.
Our data have revealed a novel interaction between Tudor-SN and ADAR1, and verified the presence of both proteins in stress granules during cell stress. In particular, these findings support the idea that ADAR1 p150 is important in various stress responses. Future studies will be key to better understanding its exact function.
MATERIALS AND METHODS
Transfections
HeLa cells were maintained in D-MEM with GlutaMAX-I (Gibco) supplemented with 10% (v/v) fetal bovine serum. 2 × 105 HeLa cells per well (six-well plate) were used for transfections. Plasmids (mCherry–TSN, mCherry–4SN, mCherry–SN1/2, mCherry–SN3/4, YFP–TSN, GFP–ADAR1 p150, GFP–ADAR2, and mCherry–ADAR1 p150) were transfected into HeLa cells using Lipofectamine-2000 (LF-2000; Invitrogen), and cells visualized after 24 h. Poly(IC) (10 ng; Sigma) was typically used to transfect HeLa cells using LF-2000, and cells were visualized after 7 h. Mock transfections used LF-2000 alone. For visualization of poly(IC) by immunofluorescence, 100 ng poly(IC) was used. For arsenite treatment, cells were incubated in media supplemented with 0.5 mM sodium arsenite (Sigma) for 30 min, then allowed to recover for 30 min in the absence of arsenite (Kedersha et al. 2002). To inhibit nuclear export, LMB (Sigma) was added (5 ng/mL) for 5 h before fixation. RNA was isolated with TRIzol, and cell lysates prepared using NET buffer. Protein concentrations were determined by Bradford assays.
Expression constructs
GFP–ADAR1 p150 and GFP–ADAR2 were as described previously (Desterro et al. 2003). Other constructs were prepared using standard cloning procedures (Sambrook and Russell 2000). For YFP–TSN, full-length Tudor-SN (amino acid residues 1–910) was amplified by PCR using primers containing BamH1 and Xba1 restriction sites, and inserted into pEYFP-C1 (Clontech). For mCherry–Tudor-SN or Tudor-SN–mCherry (mC–TSN or TSN–mC), full-length Tudor-SN (residues 1–910) was amplified by PCR using primers containing EcoR1 and BamH1 restriction sites, and inserted into pmCherry-C1 or pmCherry-N1, respectively (Clontech). The same strategy was used to clone the following Tudor-SN domains into pmCherry-C1 or pmCherry-N1: 4SN (residues 1–673), SN1/2 (residues 1–333), SN3/4 (residues 341–673), and eTD (residues 678–897). For mCherry–ADAR1 p150, full-length ADAR1 p150 (amino acid residues 1–1226) was amplified by PCR using primers containing Bcl1 and Xho1 restriction sites, and inserted into pmCherry-C1 (Clontech). All constructs were confirmed by sequencing.
Immunoprecipitations
For analysis of TSN–mC, 4SN–mC, SN1/2–mC, eTD–mC, GFP, and GFP–ADAR1 p150, recombinant proteins were expressed in HeLa cells and lysates prepared after 24 h. For immunoprecipitation of endogenous Tudor-SN or Tudor-SN–mC, 4SN–mC, SN1/2–mC, and eTD–mC, 1 mg HeLa cell lysate was diluted in 1 mL of NET buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% [v/v] NP40, 1 mM EDTA). For RNase A treatment, 100 ng RNase A was added to diluted lysates and incubated at 25°C for 20 min prior to clarification by centrifugation at 10,000g (10 min, 4°C). Agarose gel electrophoresis and RT/qPCR confirmed that the RNase treatment was efficient (data not shown). Lysates were subsequently mixed with specific antibodies or rabbit PI serum and incubated with rotation for 3 h at 4°C. Protein A Dynabeads (Invitrogen) were then added and incubated for a further 3 h. For GFP–ADAR1 p150 or GFP, 1 mg cell lysates were mixed with 30 μL of magnetic GFP–Trap (ChromoTek) in 1 mL NET buffer for 2.5 h at 4°C, with rotation. All beads were washed extensively with NET buffer and proteins were eluted using SDS buffer.
Immunofluorescence
HeLa cells were plated on 13-mm glass cover slips in six-well plates, and grown until cells reached 50%–70% confluency. Cells were washed in PBS, incubated 20 min in 4% (v/v) paraformaldehyde, and then rewashed in PBS. Cells were permeabilized in 0.5% Triton-X (Fisher Scientific) for 5 min, washed with PBS, then incubated in blocking buffer (1% [w/v] BSA in PBS) for 1 h. Cells were subsequently incubated for 1 h with primary antibodies diluted in blocking buffer, washed extensively with PBS, then incubated with secondary antibodies for 1 h. Primary antibodies used were G3BP (611127; BD Biosciences), TIAR (Sc-1749), ADAR1 (sc-73408) (Santa Cruz), Tudor-SN (Scadden 2007), and Dcp1 (Lykke-Andersen and Wagner 2005). Secondary antibodies were labeled with Rhodamine Red-X RRX (red), DyLight 488 (green), or Coumarin AMCA (blue) (Jackson ImmunoResearch). Cells were washed three times with PBS, stained with 50 μg/mL DAPI (Invitrogen) for 1 min, then mounted in ProLong Gold antifade reagent (Invitrogen). Cells were viewed and photographed using either a Zeiss Axioimager M1 microscope (Carl Zeiss MicroImaging) or a confocal Leica-SP2 UV (Leica Microsystems). The images were compiled using Adobe Photoshop.
Immunoblots
Proteins (50 μg) were separated by SDS-PAGE, transferred to PVDF, and detected using ECL. Antibodies were p-PKR (sc-16565), ADAR1 (sc-73408), HuR (sc-5261), and TIAR (sc-1749) (Santa Cruz); eIF4E (610269), eIF4G (610536), and G3BP (611127) (BD Biosciences); Dcp1 (Lykke-Andersen and Wagner 2005), RFP (ab62341) and p-eIF2α (ab4837; Abcam); and actin (A-2066; Sigma). Secondary antibodies were HRP coupled (Jackson ImmunoResearch).
RT/qPCR
DNase-treated RNA (2 μg) was reverse-transcribed (RT) using oligo (dT) and AMV-Reverse Transcriptase (Promega). –AMV-RT controls were performed. qPCR was performed using a Rotor-Gene 6000 (Corbett) and SYBR Green Jumpstart Taq ready mix (Sigma). Fold-change in mRNA was relative to control, and normalized to GapDH. Primer sequences were described previously (Vitali and Scadden 2010).
ACKNOWLEDGMENTS
We thank Patrice Vitali and Laura Hewitt for helpful comments, and Jens Lykke-Anderson (University of California, San Diego) for the Dcp1a antibody. Funding was from the BBSRC.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.027656.111.
REFERENCES
- Anderson P, Kedersha N 2002. Visibly stressed: The role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones 7: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N 2008. Stress granules: The Tao of RNA triage. Trends Biochem Sci 33: 141–150 [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N 2009. RNA granules: Post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol 10: 430–436 [DOI] [PubMed] [Google Scholar]
- Barak M, Levanon EY, Eisenberg E, Paz N, Rechavi G, Church GM, Mehr R 2009. Evidence for large diversity in the human transcriptome created by Alu RNA editing. Nucleic Acids Res 37: 6905–6915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass BL 2002. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem 71: 817–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow M, Futreal PA, Wooster R, Stratton MR 2004. A survey of RNA editing in human brain. Genome Res 14: 2379–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Parker R 2009. Eukaryotic stress granules: The ins and outs of translation. Mol Cell 36: 932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Yoon J-H, Parker R 2011. Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. J Cell Sci 124: 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callebaut I, Mornon JP 1997. The human EBNA-2 coactivator p100: Multidomain organization and relationship to the staphylococcal nuclease fold and to the tudor protein involved in Drosophila melanogaster development. Biochem J 321: 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudy AA, Ketting RF, Hammond SM, Denli AM, Bathoorn AMP, Tops BBJ, Silva JM, Myers MM, Hannon GJ, Plasterk RHA 2003. A micrococcal nuclease homologue in RNAi effector complexes. Nature 425: 411–414 [DOI] [PubMed] [Google Scholar]
- Chen L-L, Carmichael GG 2009. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: Functional role of a nuclear noncoding RNA. Mol Cell 35: 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L-L, DeCerbo JN, Carmichael GG 2008. Alu element-mediated gene silencing. EMBO J 27: 1694–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desterro JMP, Keegan LP, Lafarga M, Berciano MT, O'Connell M, Carmo-Fonseca M 2003. Dynamic association of RNA-editing enzymes with the nucleolus. J Cell Sci 116: 1805–1818 [DOI] [PubMed] [Google Scholar]
- Eckmann CR, Neunteufl A, Pfaffstetter L, Jantsch MF 2001. The human but not the Xenopus RNA-editing enzyme ADAR1 has an atypical nuclear localization signal and displays the characteristics of a shuttling protein. Mol Biol Cell 12: 1911–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Ge L, Shao J, Su C, Zhao H, Saarikettu J, Yao X, Yao Z, Silvennoinen O, Yang J 2010. Tudor-SN interacts with and co-localizes with G3BP in stress granules under stress conditions. FEBS Lett 584: 3525–3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George CX, Samuel CE 1999. Characterization of the 5′-flanking region of the human RNA-specific adenosine deaminase ADAR1 gene and identification of an interferon-inducible ADAR1 promoter. Gene 229: 203–213 [DOI] [PubMed] [Google Scholar]
- Hartner JC, Walkley CR, Lu J, Orkin SH 2009. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat Immunol 10: 109–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH 2000. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406: 78–81 [DOI] [PubMed] [Google Scholar]
- Hundley HA, Krauchuk AA, Bass BL 2008. C. elegans and H. sapiens mRNAs with edited 3′ UTRs are present on polysomes. RNA 14: 2050–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Chen S, Gilks N, Li W, Miller IJ, Stahl J, Anderson P 2002. Evidence that ternary complex (eIF2-GTP-tRNAiMet)-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol Biol Cell 13: 195–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol 169: 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo N, Wolff B, Sekimoto T, Schreiner EP, Yoneda Y, Yanagida M, Horinouchi S, Yoshida M 1998. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp Cell Res 242: 540–547 [DOI] [PubMed] [Google Scholar]
- Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. 2004. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol 22: 1001–1005 [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J, Wagner E 2005. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev 19: 351–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse DP, Aruscavage PJ, Bass BL 2002. RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proc Natl Acad Sci 99: 7906–7911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paukku K, Yang J, Silvennoinen O 2003. Tudor and nuclease-like domains containing protein p100 function as coactivators for signal transducer and activator of transcription 5. Mol Endocrinol 17: 1805–1814 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW 2000. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Scadden ADJ 2005. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat Struct Mol Biol 12: 489–496 [DOI] [PubMed] [Google Scholar]
- Scadden ADJ 2007. Inosine-containing dsRNA binds a stress-granule-like complex and downregulates gene expression in trans. Mol Cell 28: 491–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scadden ADJ, Smith CWJ 2001. Specific cleavage of hyper-edited dsRNAs. EMBO J 20: 4243–4252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra MJ, Smolter PE, Westhof E 2004. Pronounced instability of tandem IU base pairs in RNA. Nucleic Acids Res 32: 1824–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist CJA, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, Bairoch A, Hulo N 2010. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res (Suppl 1) 38: D161–D166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark GR, Kerr IM, Williams BRG, Silverman RH, Schreiber RD 1998. How cells respond to interferons. Annu Rev Biochem 67: 227–264 [DOI] [PubMed] [Google Scholar]
- Strehblow A, Hallegger M, Jantsch MF 2002. Nucleocytoplasmic distribution of human RNA-editing enzyme ADAR1 is modulated by double-stranded RNA-binding domains, a leucine-rich export signal, and a putative dimerization domain. Mol Biol Cell 13: 3822–3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth AM, Li Z, Cattaneo R, Samuel CE 2009. RNA-specific adenosine deaminase ADAR1 suppresses measles virus-induced apoptosis and activation of protein kinase PKR. J Biol Chem 284: 29350–29356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourriere H, Gallouzi I-e, Chebli K, Capony JP, Mouaikel J, van der Geer P, Tazi J 2001. RasGAP-associated endoribonuclease G3BP: Selective RNA degradation and phosphorylation-dependent localization. Mol Cell Biol 21: 7747–7760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente L, Nishikura K, Kivie M 2005. ADAR gene family and A-to-I RNA editing: Diverse roles in posttranscriptional gene regulation. In Progress in nucleic acid research and molecular biology (ed. K Moldave), Vol. 79, pp. 299–338. Academic Press, London [Google Scholar]
- Vitali P, Scadden ADJ 2010. Double-stranded RNAs containing multiple IU pairs are sufficient to suppress interferon induction and apoptosis. Nat Struct Mol Biol 17: 1043–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QD, Miyakoda M, Yang WD, Khillan J, Stachura DL, Weiss MJ, Nishikura K 2004. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J Biol Chem 279: 4952–4961 [DOI] [PubMed] [Google Scholar]
- Yang J, Aittomaki S, Pesu M, Carter K, Saarinen J, Kalkkinen N, Kieff E, Silvennoinen O 2002. Identification of p100 as a coactivator for STAT6 that bridges STAT6 with RNA polymerase II. EMBO J 21: 4950–4958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Valineva T, Hong J, Bu T, Yao Z, Jensen ON, Frilander MJ, Silvennoinen O 2007. Transcriptional co-activator protein p100 interacts with snRNP proteins and facilitates the assembly of the spliceosome. Nucleic Acids Res 35: 4485–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]