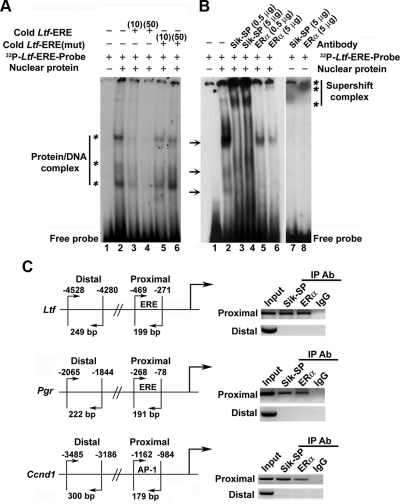
Fig. 3.
Analysis of E2-dependent recruitment of Sik-SP in conjunction with ERα to gene-specific promoter. A, EMSA for Sik-SP recruitment to ERE via ERα binding. Primer sequence designed from half consensus ERE of mouse Ltf gene was used: 5′-TGTCACAGGTCAAGGTAACCCACAAAT-3′. The mutant (mut) primer was designed after randomization of the above sequence. 32P-end-labeled probes were generated by kinase reaction using [γ32P]ATP and T4 polynucleotide kinase. The double stranded primers were prepared after heating to 90 C for 2–3 min, followed by slow cooling. Binding of E2-treated uterine nuclear proteins to ERE probes was achieved as described in Materials and Methods. For competition assays, cold or mut primers were added at 10- or 50-fold molar excess. Samples were loaded onto 5% nondenaturing polyacrylamide gel and run at 150 V for 2.5 h. The gel was dried and exposed to x-ray film (Kodak) at −80 C. Bands as shown by asterisks on the left indicate specific shifts due to formation of protein-DNA complexes. B, Supershift assays were performed by adding Sik-SP or ERα antibody at indicated concentrations to the above reaction. Supershift bands are indicated by asterisks on the right. Arrows denote the position of bands that are abolished after the addition of either type of antibodies, suggesting both Sik-SP and ERα recruit as a complex to the ERE. C, ChIP analysis of Sik-SP and ERα recruitment to E2-responsive Ltf, Pgr, and Ccnd1 genes. ChIP analysis was performed using Sik-SP or ERα antibody (Ab) or normal rabbit serum (as IgG control) as described by us (47). Primers designed for the proximal and distal regions were previously described by us (3). The presence of the promoter DNA before immunoprecipitation (IP) was confirmed by PCR (input).