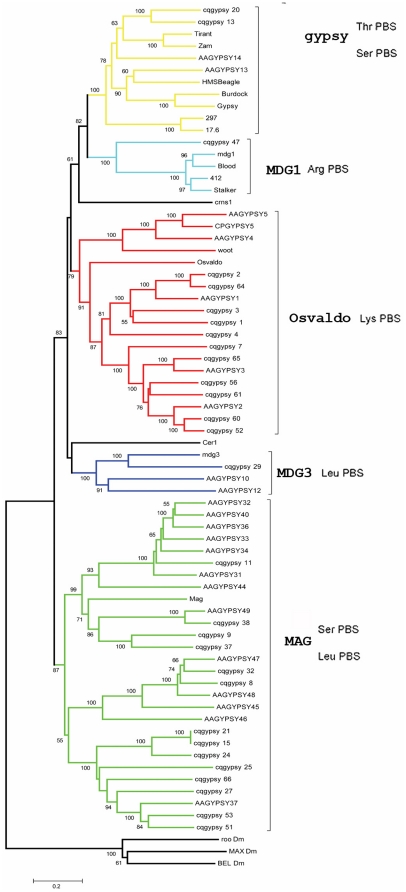
Figure 1. Evolutionary relationships of C. quinquefasciatus LTR-retrotransposons.
Phylogenetic relationships of the LTR retrotransposons based on the amino acids alignment of the conserved RT, RNase H and INT domains. The clades in which fall retrotransposons detected in this paper are indicated with different colors, along with the most common tRNA complementary to the PBS is indicated for each homogeneous group. Elements from this study are indicated as “cpgypsy_” followed by a number. AAGYPSY# elements are LTR retrotransposons identified in previous analyses [17]. The N-J bootstrap values supporting the internal branches are indicated at the nodes. Only bootstrap values greater than 50% are reported. Bel-like elements were used as outgroup. Note that, for families composed of two or more copies (see table 1), representative elements (see file S1) were used for the phylogenetic analyses.