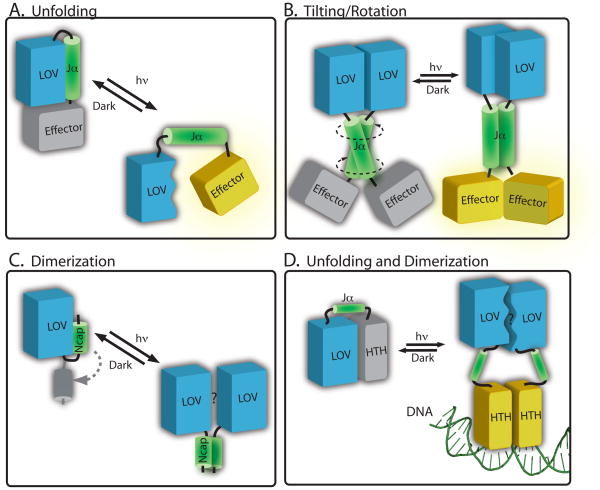
Figure 6.
Structural models of signalling in LOV proteins (A) In phototropin-type signaling, cysteinyl-flavin adduct formation induces conformational change in the LOV2 domain (blue) that results in disruption of the interaction with the Jα helix (green). This leads to effector domain activation (yellow). Data from YtvA and bacterial LOV histidine kinases evidence models in which illumination of a LOV domain (blue) can induce conformational changes in an extended Jα helix (green), causing (B) tilting or rotational motion to activate the effector (in yellow). (C) In VVD, light activation leads to rearrangement of the N-terminal cap (in green) and a subsequent change in protein dimerization. (D) In the E. litoralis LOV-HTH protein, EL222, it has been proposed that light activation disrupts the interaction surface between the LOV domain and the HTH domain. This light-driven structural change leads to dimerization of the protein on DNA106.