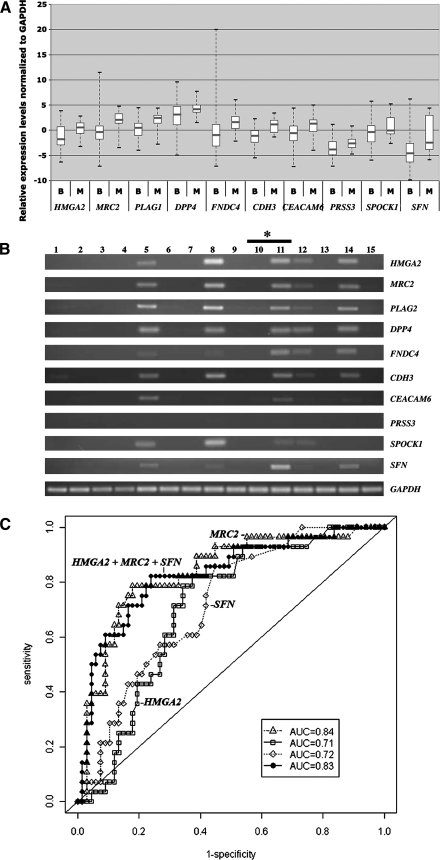
FIG. 2.
QRT-PCR analysis of HMGA2, MRC2, PLAG1, DPP4, FNDC4, CDH3, CEACAM6, PRSS3, SPOCK1, and SFN. (A) Relative gene expression levels normalized to GAPDH in 67 benign (B) and 28 malignant (M) FNAs were determined using gene-specific primers as described in Materials and Methods. The upper and lower limits of each box represent ‘‘third’’and ‘‘first’’quartiles, respectively. Gray line, medians; whiskers, extreme measurements. Note: as expected from our previous microarray analysis (19), all 10 genes appeared to be overexpressed in malignant FNAs compared with benign samples. (B) The agarose gel electrophoretic analysis representing the mRNA expression in 15 intraoperative needle aspirates, including seven adenomatoid nodules (lanes 1, 2, 4, 7, 10, 13, 15), one follicular adenoma (lane 6) and seven-papillary thyroid carcinoma (lanes 3, 5, 8, 9, 11, 12, 14). GAPDH expression served as a loading control. *The RNA samples in lanes 10 and 11 are isolated from two different nodules/tumor, but from the same patient. (C) ROC analysis of mRNA expression (QRT-PCR) in 95 intraoperative FNA samples. Results are shown for the AUC for one-gene [HMGA2 (□; AUC=0.71), MRC2 (▵; AUC=0.84), SFN (◊; AUC=0.72)], and three-gene (HMGA2+MRC2+SFN [•; AUC=0.83]) combinations. The solid diagonal line denotes an AUC=0.50. FNA, fine needle aspiration; QRT-PCR, quantitative real-time RT-PCR.