Fig. 1.
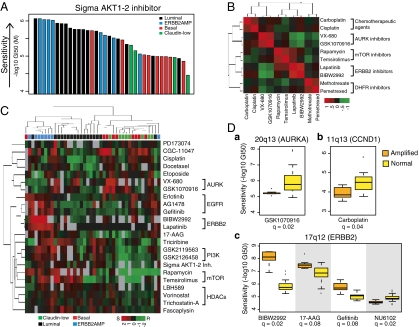
Cell lines show a broad range of responses to therapeutic compounds. (A) Luminal and ERBB2AMP cell lines respond preferentially to AKT inhibition. Each bar represents the response of a single breast cancer cell line to the Sigma AKT1-2 inhibitor and is colored according to subtype. Cell lines are ordered by decreasing sensitivity [−log10(GI50)]. (B) Drug-response profiles for compounds with similar mechanisms and targets are highly correlated. Heatmap shows hierarchical clustering of pairwise correlations between responses of breast cancer cell lines treated with one of eight compounds. Red indicates positively correlated sensitivity across the panel of cell lines. Green indicates anticorrelated drug-response profiles. (C) Many compounds are significantly associated with subtype. Each column represents one cell line, and each row represents the median-centered −log10(GI50) for a particular compound. Both rows and columns are clustered hierarchically. Red represents sensitivity, green represents resistance, and gray represents missing values. Colored boxes below the dendogram identify sample subtype. Overall, cell lines of similar subtype tend to cluster together, as do compounds with similar targets or mechanisms. (D) CNAs are associated with compound response. Boxplots show distribution of response sensitivity for cell lines with amplified (A) and normal (N) copy number at the noted genomic locus. (a) 20q13 (STK15/AURKA) amplification is associated with GSK1070916 response (A = 7; N = 26 samples). (b) Amplification at 11q13 (CCND1) is associated with response to carboplatin (A = 9; N = 28 samples). (c) 17q12 (ERBB2) amplification is associated with sensitivity to BIBW2992 (A = 6; N = 19 samples), 17-AAG (A = 7; N = 27 samples), and gefitinib (A = 7; N = 18 samples), as well as resistance to NU6102 (A = 6; N = 21 samples).