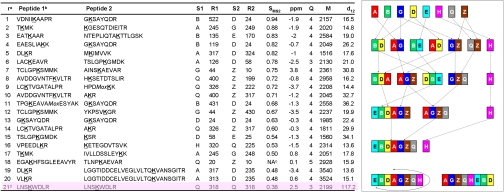
Fig. 2.
Twenty-one identified cross-links between subunits that have associated distances. The specific cross-linked lysine residues are listed together with their MS parameters. Cross-links that involved Lys residues not seen in the X-ray structure of the 1Q3R template are omitted. The first 18 entries are used for the combinatorial analysis (Fig. 3). Mox is oxidized methionine. aThe column headers are r for the rank in this list, S1 for the name of the first subunit in the cross-link, R1 for the lysine residue in S1, S2 and R2 for the name and position of the second subunit, SMS2 is the MS/MS fit score (see text), ppm is the mass error of the precursor ion in parts per million, Q is the ion charge, M is the molecular mass of the ion rounded to the nearest dalton, d12 is the distance between the Cα atoms of the cross-linked lysine residues R1 and R2 in the OMS model of TRiC (Fig. 4). bThe most likely cross-linked lysine is underlined in Peptide 1 and Peptide 2. cThis one cross-link was included in spite of its low MS/MS fit score (0.32) as there are at least four identified MS/MS fragmentation sites on each peptide. dThe last cross-link marked in pink highlight is not used in any analysis. The inset on the right shows that the six most reliable unique cross-links between subunit pairs: BD, AG, BE, AD, GZ and QZ assemble to ring arrangement EBDAGZQH, the correct arrangement (after rotation), without need for any modeling. Modeling we do here is essential to show this arrangement is unique and statistically significant.