Abstract
In Escherichia coli, the replication origin oriC consists of two functional regions: the duplex unwinding element (DUE) and its flanking DnaA-assembly region (DAR). ATP-DnaA molecules multimerize on DAR, unwinding DUE for DnaB helicase loading. However, DUE-unwinding mechanisms and functional structures in DnaA–oriC complexes supporting those remain unclear. Here, using various in vitro reconstituted systems, we identify functionally distinct DnaA sub-complexes formed on DAR and reveal novel mechanisms in DUE unwinding. The DUE-flanking left-half DAR carrying high-affinity DnaA box R1 and the ATP-DnaA-preferential DnaA box R5, τ1-2 and I1-2 sites formed a DnaA sub-complex competent in DUE unwinding and ssDUE binding, thereby supporting basal DnaB loading activity. This sub-complex is further subdivided into two; the DUE-distal DnaA sub-complex formed on the ATP–DnaA-preferential sites binds ssDUE. Notably, the DUE-flanking, DnaA box R1–DnaA sub-complex recruits DUE to the DUE-distal DnaA sub-complex in concert with a DNA-bending nucleoid protein IHF, thereby promoting DUE unwinding and binding of ssDUE. The right-half DAR–DnaA sub-complex stimulated DnaB loading, consistent with in vivo analyses. Similar features are seen in DUE unwinding of the hyperthermophile, Thermotoga maritima, indicating evolutional conservation of those mechanisms.
INTRODUCTION
Localized DNA-duplex unwinding is a fundamental event during the initiation of chromosomal replication. This event is tightly regulated by an initiation complex, a highly ordered nucleoprotein complex (which includes AAA+ family proteins) that is conserved in all domains of life (1–3). In Escherichia coli, DnaA, a AAA+ protein, forms a specific multimer with the replication origin, oriC, to yield the initiation complex (4–6). In this complex, the DnaA-binding protein DiaA stabilizes DnaA multimers (7,8).
Escherichia coli oriC carries two functional regions: an AT-rich duplex unwinding element (DUE) [including an AT-cluster, and AT-rich 13-mer repeats termed L (left), M (middle) and R (right)] and a flanking DnaA assembly region (DAR) (including DnaA boxes R1–5, I1-3 and τ1–2) (Figure 1A) (5,6,9). Unlike ADP–DnaA–oriC complexes, ATP–DnaA–oriC complexes specifically unwind the DUE in the presence of superhelical tension, resulting in the open complex (10–12). The T-rich strand, but not the A-rich strand, of the resultant single-stranded (ss)DUE binds to an ATP–DnaA oligomer, which stabilizes the open complex (13–15). A cluster of T within ssDUE is indicated to be crucial in the DnaA binding (13,16). DnaB helicases are then loaded onto the ssDUE with the aid of both DnaA and the helicase-loader, DnaC. This is followed by the sequential loading of DnaG primase and the DNA polymerase III holoenzyme (4,17,18).
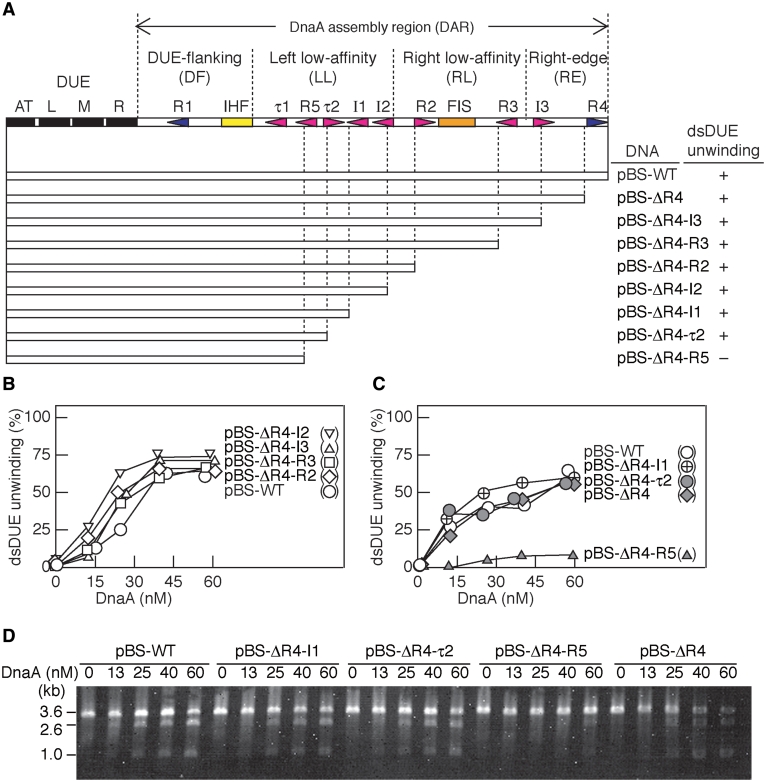
Figure 1.
Proposed E. coli oriC sub-structures and determination of the minimum region required for DUE unwinding in the plasmid oriC. (A) The overall structure of oriC and its sub-structures proposed in this study are shown at the top of the panel. The AT-rich motifs within the DUE, IHF-binding site and Fis-binding site are indicated by black, yellow and orange bars, respectively. DnaA binding motifs are indicated by arrowheads (blue for high-affinity sites R1 and R4 and red for low-affinity sites). The proposed DAR substructures are indicated above of the overall structure. The oriC regions used for deletion analysis (open bars) are shown below the oriC structure. The supercoiled form of pBSoriC, or its derivatives (3.4 nM), ATP-DnaA (0–60 nM) and IHF (55 nM) were used for the P1 nuclease assay. The unwinding activity at 60 nM ATP–DnaA is shown below the heading ‘dsDUE unwinding’. +, wild-type level; –, inactive. (B–D) The number of P1 nuclease-digested oriC DNA molecules per that of input DNA was analyzed by 1% agarose gel electrophoresis and ethidium bromide staining and shown as a percentage [dsDUE unwinding (%)] (B and C). The gel images are shown in panel D and Supplementary Figure S1.
DnaA comprises four distinct domains (6). Domain I interacts with multiple proteins including DnaB, DiaA and the DnaA domain I within another protomer in the DnaA complexes (7,8,19,20). Domain II is a flexible linker (19,21). Domain III contains motifs characteristic of the AAA+ family (e.g. Walker A/B, Sensor 1/2 and an Arg-finger) (22,23). Domain IV is a DNA-binding domain with a helix–turn–helix motif (22,24).
Notably, the Arg-finger (Arg285) and B/H motifs (Val211 and Arg245) within domain III play crucial and specific roles in ATP-dependent activation of the initiation complexes (13,25). DnaA Arg285 is required for inter-ATP–DnaA interactions and co-operative binding to low-affinity DnaA-binding sites within the oriC DAR (25). Similar to typical AAA+ proteins, the ATP–DnaA multimers on oriC likely adopt a spiral structure containing a central pore, and the DnaA Arg285 Arg-finger interacts with ATP bound to the adjacent protomer (25,26). Moreover, our recent analysis reveals that unlike oriC complexes formed with wild-type ATP–DnaA, those with ATP–DnaA V211A or R245A are specifically defective in the ssDUE binding, although these mutants preserve the Arg-finger-mediated inter-DnaA interaction (13). Hydrophobic or basic moiety of the residue corresponding to the Val-211 or Arg-245 is highly conserved in DnaA homologs and thus these residues are named H and B motifs, respectively. Structural analyses suggest that within the proposed DnaA multimer spiral, the B/H motifs of each protomer are regularly spaced on the pore surface, which likely supports direct and stable binding to ssDUE (13).
ATP– or ADP–DnaA binds with high affinity to DnaA boxes R1 and R4, which reside at the both edges of DAR (27) (Figure 1A). A moderate-affinity site (R2) and low-affinity sites (R3 and R5, I1-3 and τ1-2) located between R1 and R4 boxes preferentially bind to ATP–DnaA oligomers, rather than to ADP–DnaA (25,28). The formation of ATP–DnaA oligomers at these moderate- and low-affinity sites is important for the activation of DnaA–oriC complexes in DUE unwinding. Also, DAR contains IHF- and FIS-binding sites (Figure 1A). IHF and FIS are nucleoid-associated proteins, and often called histone-like proteins (29). IHF binding to the IHF-binding site stimulates replication initiation (30–34); Mutations in the IHF-binding site decrease the initiation activity of mini-chromosomes and the chromosomal oriC in vivo. FIS binding and dissociation support the initiation of replication at a specific time during the cell cycle (31,34,35).
As described above, an ATP–DnaA–oriC complex is multifunctional and promotes stepwise reactions of DUE unwinding, stabilization of the unwound ssDUE and DnaB loading. Based on the fact that the Arg-finger and B/H motifs of DnaA are required by only a certain sub-group(s) of DnaA protomers, we previously suggested that the ATP–DnaA–oriC complex comprises several sub-groups of DnaA protomers, each associated with distinct function (13,25). However, the organization and specific functions of the DnaA sub-complexes remains unclear.
In this study, to reveal the fundamental mechanisms involved in origin unwinding and helicase loading, we analyze DnaA sub-complexes formed on a set of truncated oriC fragments and specific activities of those using in vitro reconstituted systems. Based on the results, we subdivided the DAR region into four (Figure 1A): the DUE-flanking (DF), left low-affinity (LL), right low-affinity (RL) and right-edge (RE) regions (Figure 1A). Particularly, the DF region carries high-affinity DnaA box R1 and the LL region carries ATP–DnaA-preferential binding sites DnaA box R5 and τ1-2. A DnaA sub-complex containing the DUE, DF and LL regions was fully active in DUE unwinding and ssDUE binding, which supports DnaB loading basically. In this complex, a DnaA sub-complex formed on the LL region bind ssDUE, which is specifically promoted by the DF region-bound DnaA in a cooperative manner, thereby overall providing structural basis of ssDUE-binding complex. This process was enhanced by binding of a DNA-bending protein IHF to the specific sites between the DnaA-binding sites within the DF and LL regions. DnaA sub-complexes formed on the RL and RE regions stimulate DnaA multimerization and DnaB loading. These results are consistent with those of previous in vivo analyses. Similar results were obtained using DnaA ortholog and the cognate oriC from the ancient hyperthermophilic eubacterium Thermotoga maritima, suggesting that the mechanisms we have identified are highly conserved in eubacterial species.
MATERIALS AND METHODS
Proteins, DNA and buffers
DnaA proteins were purified as previously described (13,36). IHF was overexpressed in MC1061 cells and purified as previously described (37). DNA, plasmids and the buffers used are described in Supplementary Data.
DUE unwinding assay
The assay was performed as previously described (13). Briefly, DnaA was incubated for 3 min at 38°C in buffer P (20 µl) containing IHF (55 nM) and a supercoiled form of pBSoriC, or its mutant derivative (4 nM), followed by incubation for 200 s at the same temperature in the presence of P1 nuclease (4 U, Yamasa Co.). After reactions were terminated by addition of 0.5% sodium dodecyl sulfate (SDS), DNA was extracted with phenol–chloroform and precipitated with ethanol, and a portion (1/10 vol) was digested with AlwNI, followed by electrophoresis in a 1% agarose gel and ethidium bromide staining.
ssDUE digestion assay using linearized oriC fragments
pOZoriEC3 derivatives were digested with HincII and EcoRI, yielding fragments of the vector DNA and ori-EcoRI. The protruding end generated by EcoRI was filled in using the DNA polymerase I large fragment and 32 P-dNTPs. The labeled DNA fragments (1.3 nM) and the ATP or ADP forms of DnaA (0–80 nM) were incubated for 5 min at 38°C in buffer P (10 µl) including IHF (0–55 nM), followed by incubation with P1 nuclease (50 mU) for 2 min. ATP– and ADP–DnaA were prepared as previously described (13). The DNA was purified using phenol–chloroform extraction. A portion (2/3 vol) was analyzed by electrophoresis in a 6% polyacrylamide gel. Radioactivity was analyzed using a BAS-2500 (Fujifilm).
ssDUE binding analysis by electrophoretic mobility shift assay
This assay was performed essentially as previously described (13). ATP–DnaA and 2.5 nM of 32P-labeled ssDNA (ssDUE, ssDUE-R1 or ssDUEnon) were incubated for 10 min at 30°C in buffer G (10 µl) containing 4 µg/ml poly(dA–dT)–(dA–dT), and 4 µg/ml poly(dI–dC)–(dI–dC) in the presence of DAR derivatives (5 nM), followed by 4% polyacrylamide gel electrophoresis (PAGE) at room temperature. When tmaDnaA was used, the ATP or ADP forms of tmaDnaA were incubated for 10 min at 48°C in buffer G (10 µl) containing tmaDAR derivatives (10 nM) and 2 nM of 32P -end-labeled DNA (tma-ssDUE, ssDUEbox1, ssDUEnon1 or dsDUEbox1).
DNase I footprint
The end-labeled tma-oriC fragment (10 nM) and tmaDnaA (0–800 nM) were incubated for 10 min at 48°C in buffer G (10 µl) containing 5 mM calcium acetate and 3 mM ATP or ADP, followed by incubation with DNase I (0.83 mU) for 4 min at the same temperature. DNA samples were analyzed by sequencing gel electrophoresis as described previously (13).
Form I* formation assay
This assay was performed essentially as previously described (7) (see Supplementary Data).
Pull-down assay
This assay was performed essentially as described previously (7). The biotinylated oriC DNA derivative or a control DNA was incubated at 4°C for 10 min in buffer G (10 µl) containing DnaA (1 µM), HisDnaB (0.5 µM), DnaC (0.5 µM) and λ phage DNA (75 ng). Biotinylated DNA-bound materials were recovered using streptavidin-coated beads (Promega), washed in buffer G′ (12.5 µl) including 12 mM ammonium sulfate, dissolved in SDS sample buffer and analyzed by SDS-11% PAGE and silver staining. The biotinylated DNA remained in the streptavidin-unbound fraction was extracted with phenol–chloroform, precipitated with glycogen in ethanol and analyzed quantitatively by 6% PAGE and ethidium bromide staining. In parallel, quantitative control DNA was used for ethanol precipitation and electrophoresis. The amounts of the isolated DNA were deduced using those of the control DNA and were used to deduce the amounts of beads-bound DNA. Fifty to sixty percent of the input DNA were recovered in the beads-bound fraction in these experiments.
RESULTS
The R1–R5 region in DAR is sufficient for DUE unwinding of supercoiled oriC
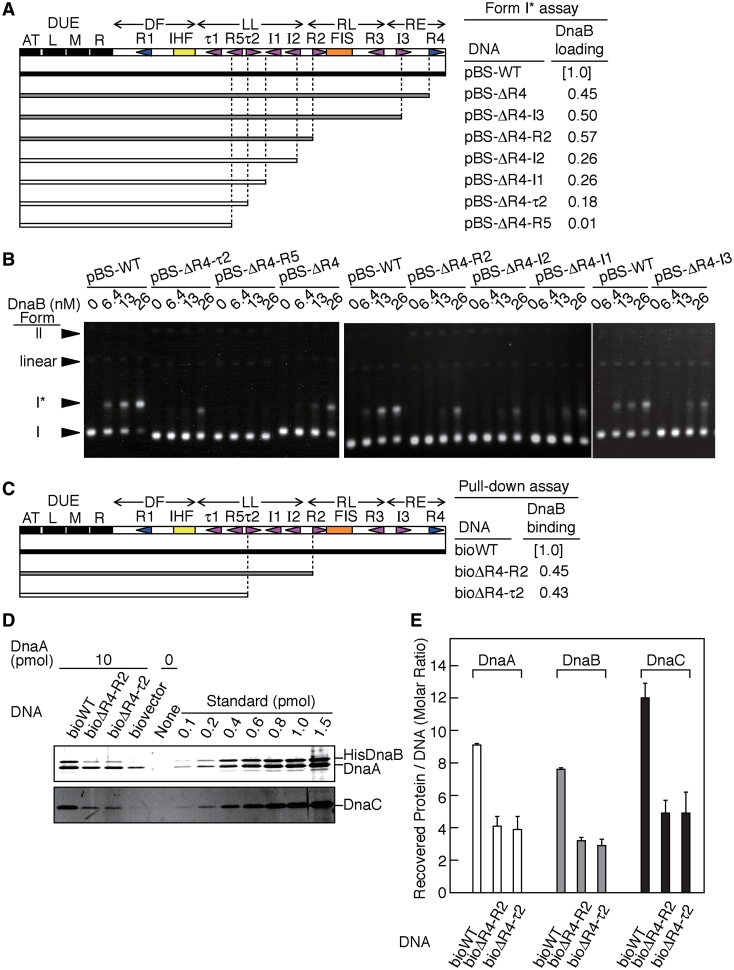
To identify the minimal requirements within the oriC DAR for DUE unwinding, we performed a P1 nuclease assay using plasmids containing serial deletions from the right end of oriC (Figure 1A, Supplementary Figure S1). In this assay, ATP–DnaA (in the presence of IHF) unwinds the DUE duplex of a supercoiled form of the wild-type oriC plasmid, pBSoriC (pBS-WT), or its derivatives (13,30). The resulting ssDNA is susceptible to cleavage by P1 nuclease. The results showed that all the mutant pBSoriC derivatives, except for pBS-ΔR4-R5, retained the ability to unwind DUE at a level comparable with that of pBS-WT (Figure 1B and C, Supplementary Figure S1). In addition, the initial rate of DUE unwinding was similar to between pBS-WT and pBS-ΔR4-τ2 (Supplementary Figure S1). These indicate that pBS-ΔR4-τ2 contained the minimal DAR region required for DUE unwinding in vitro. The oriC region of pBS-ΔR4-τ2 contains the DUE, an IHF-binding site, the high-affinity DnaA box R1 and the LL regions τ1 and R5 (Figure 1A).
ssDUE binding to ATP–DnaA multimers on DAR can be analyzed using linear oriC
When the oriC region is located on a circular form DNA, DUE unwinding requires superhelicity (10,30). Also, the upper strand of the ssDUE containing M and R motifs is demonstrated to bind to ATP–DnaA multimers on oriC in a manner dependent on DnaA H-motif Val211 and B-motif Arg245 within the AAA+ domain, resulting in stabilization of the unwound ssDUE state (13). As such, DUE unwinding is sustained by torsional stress of supercoiled oriC DNA and ssDUE binding of DnaA multimers.
To eliminate the effect of torsional stress and to specifically analyze the effect of ssDUE binding by DnaA multimers in DUE unwinding, we developed a novel assay using linearized oriC DNA (Figure 2). The DUE, located at the end of the linear DNA, transiently unwinds at 38°C by thermal denaturation of the duplex DNA end (Figure 2A). If the resultant ssDUE forms stable complexes with ATP–DnaA multimers, then the sensitivity of the DNA to P1 nuclease should increase. As the AT-rich M and R motifs within DUE are sufficient for interaction with ATP-DnaA multimers (13,14), we first constructed a 32 P-end-labeled oriC fragment (ori-EcoRI) bearing these motifs near the DNA terminus (Figure 2A). A vector plasmid-derived fragment without the oriC region (vector) was similarly end-labeled and used as an internal control. These DNA fragments were then incubated with ATP–DnaA and P1 nuclease. The ori-EcoRI DNA (EC3-WT), but not the vector DNA, was digested in a manner dependent on ATP–DnaA (Figure 2B and C), P1 nuclease and incubation at 38°C (Supplementary Figure S2). The efficiency of the digestion was comparable to that seen for DUE unwinding of supercoiled pBSoriC (Figure 1).
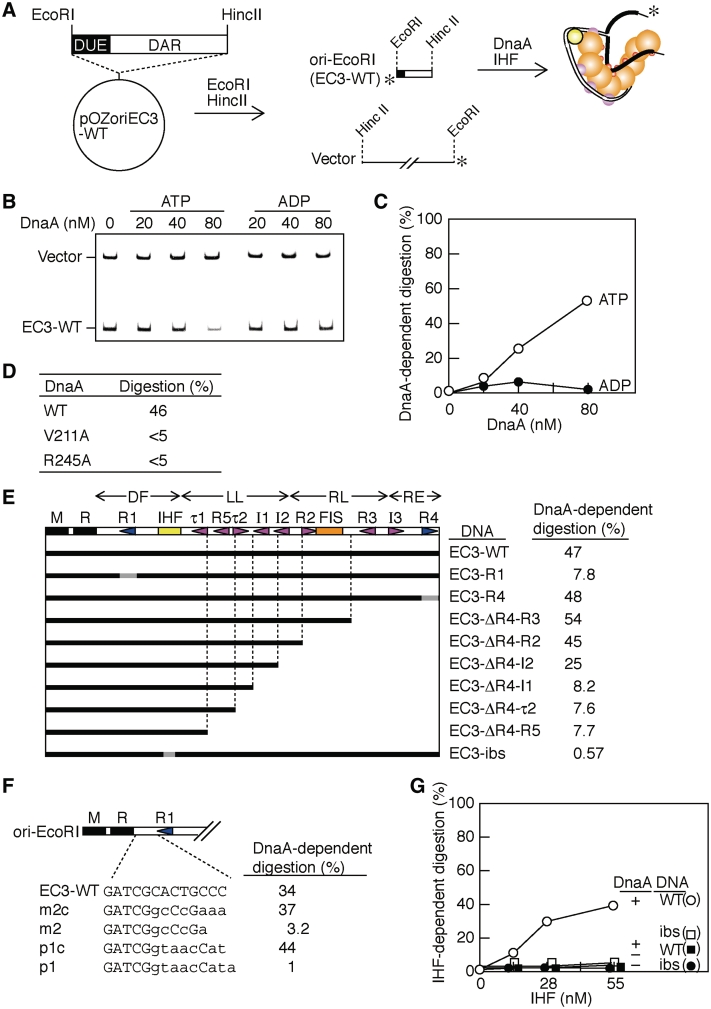
Figure 2.
ssDUE digestion assay using oriC fragments for analysis of DnaA–DAR complexes. (A) The plasmid, pOZoriCEC3, was digested using HincII and EcoRI, yielding DNA fragments of vector DNA (vector) and ori-EcoRI (EC3-WT). These fragments were end-labeled and incubated with ATP–DnaA and IHF, followed by analysis using P1 nuclease and polyacrylamide gel electrophoresis. The postulated structure of the DnaA multimers complexed with ori-EcoRI is also illustrated. An alternative structural model is also conceivable but it is not shown here for the simplicity (see ‘Discussion’ section). The DnaA domains III and IV and IHF are indicated by orange, pink and yellow balls, respectively. The DnaA B/H-motifs within domain III are indicated by small red balls. For simplicity, DnaA domains I–II are not shown. The DUE region is indicated by thick black bars. (B and C) The P1 nuclease sensitivity of the ori-EcoRI (EC3-WT) (1.3 nM) was analyzed in the presence of IHF (55 nM) and the indicated concentrations of either ATP–DnaA (ATP) or ADP–DnaA (ADP). Reaction mixtures were incubated for 5 min at 38°C, followed by incubation with P1 nuclease and polyacrylamide gel electrophoresis. A polyacrylamide gel containing the reaction products is shown (B). The ori-EcoRI (EC3-WT) and the vector remained were quantified, and the relative numbers of the remaining molecules of ori-EcoRI to that of the vector were deduced. The number obtained in the absence of DnaA was defined as 100% and the relative numbers are plotted as DnaA-dependent digestion (C). (D) The P1 nuclease sensitivity of ori-EcoRI (EC3-WT) was analyzed as above using 80 nM of wild-type ATP–DnaA (WT), ATP–DnaA V211A (V211A) or ATP–DnaA R245A (R245A) in the presence of IHF (55 nM). (E and F) The mutant derivatives of pOZoriEC3 were constructed and digested with EcoRI and HincII. The resultant ori-EcoRI derivatives are shown using black and gray bars, which indicate regions bearing wild-type sequences and base substitutions, respectively (E). In (F), only the DNA constructs between the DUE M and DnaA box R1 of ori-EcoRI derivatives are shown. The wild-type sequences and base substitutions are indicated by upper- and lower case, respectively. In (E) and (F), those fragments (1.3 nM) and vector fragments were incubated as above in the presence of IHF (55 nM) and ATP–DnaA (80 nM), followed by incubation with P1 nuclease. Relative levels in DnaA-dependent digestion of EC3-WT derivatives were deduced as above (see Supplementary Figure S2 for details). (G) EC3-WT (WT) and EC3-ibs (ibs) were incubated as above with the indicated concentrations of IHF in the presence (+) or absence (−) of ATP–DnaA (80 nM), followed by incubation with P1 nuclease. The relative molecular number of remaining ori-EcoRI was quantified as above, and that obtained in the absence of both DnaA and IHF was defined as 100%. This was used to deduce the relative levels of IHF-dependent digestion (shown as percentages, ‘IHF-dependent digestion’).
Moreover, neither the wild-type ADP–DnaA (Figure 2B and C) nor the ATP-forms of the B/H-motif mutants (V211A or R245A) (Figure 2D) promoted digestion of EC3-WT by P1 nuclease, a result consistent with that reported in our previous study (13). All these observations are consistent with the ideas that ATP–DnaA molecules form a complex with DAR and then bind ssDUE generated by thermal denaturation of the duplex in a manner independent of superhelicity and that the resulting complex stabilizes the unwound form and promotes sensitivity of ssDUE to P1 nuclease.
The DF and LL regions of DAR are crucial in ssDUE binding
Using this assay, we determined the functional substructures within the DAR involved in ssDUE binding (Figure 2E and Supplementary Figure S2). Digestion of the ssDUE was severely inhibited when DnaA box R1 was substituted with a non-sense sequence (EC3-R1; Figure 2E), indicating an important role for DnaA box R1 in ssDUE binding. Eight percent residual activity of EC3-R1 may suggest that another sub-complex formed outside of the R1 box has a considerable activity in ssDUE binding, which is effectively enhanced by DnaA–R1 complex.
Deletion of DnaA boxes R2–4 and the I3 site did not result in loss of function in this assay (EC3-R4, ΔR4-R3 and ΔR4-R2; Figure 2E). However, deletion of I2 moderately inhibited ssDUE digestion and additional serial deletions of the I1, τ2 and R5 sites severely inhibited ssDUE digestion with only 0.6–8% residual activity (EC3-ΔR4-I2, ΔR4-I1, ΔR4-τ2 and ΔR4-R5; Figure 2E). These results indicate that both the DF and LL regions, but not the RL and RE regions, are required for the full activity of ssDUE digestion, suggesting that the ATP–DnaA multimers formed on the DF and LL regions are fully active in ssDUE binding. Because the oriC region between the DUE and the R5 box was sufficient for DUE unwinding when the supercoiled form of the oriC plasmid was used (Figure 1), ATP–DnaA molecules bound to the τ2 and I1-2 sites within the LL region could act to assist ssDUE binding in the absence of torsional stress.
We further investigated if the spacing between DUE and DnaA box R1 is important for the ori-EcoRI DNA digestion. A previous report using deletion, insertion and substitution mutations within oriC indicates that the strict spacing, but not nucleotide sequence, between the DUE R motif and DnaA box R1 on oriC is required for replication initiation both in vivo and in vitro (38). Consistently, our results revealed that even a 1-bp insertion or a 2-bp deletion between the 13-mer R motif and DnaA box R1 inhibits the specific digestion of the ori-EcoRI DNA (Figure 2F). These results support the idea that the strict spacing between DUE and DnaA box R1 is crucial for the ssDUE binding.
IHF is crucial in ssDUE binding
DUE unwinding is facilitated by IHF that promotes a sharp bend of DNA (29,30). However, it is unclear whether this is related to ssDUE binding of ATP–DnaA or modulation in torsional stress of supercoiled oriC DNA. Using ori-EcoRI DNA (EC3-WT) and P1 nuclease, we demonstrated that ssDUE digestion depends on the presence of both DnaA and IHF (Figure 2G), indicating that stimulation of dsDUE unwinding requires both. Consistently, ssDUE digestion was severely inhibited when an ori-EcoRI derivative containing base substitutions (ATCAAC to AGATCG) defective in IHF binding (31) was used (Figure 2G). These indicate that IHF binding to the IHF-binding site plays a key role in ssDUE binding.
Based on these results, we propose a mechanism (termed ssDUE recruitment) that involves IHF-induced DNA bending within the DF region. This can stabilize the unwound form of DUE by promoting binding between the ssDUE and the DnaA–DAR LL sub-complex (see later).
The DnaA box R1 is dispensable in ssDUE binding
In the above experiments, ori-EcoRI carried the DUE and the DAR in cis. To further investigate the role played by the DAR in ssDUE binding, we analyzed the interaction between the ATP–DnaA–DAR complex and the ssDUE (this time added in trans) using electrophoretic mobility shift assay (EMSA) in the absence of IHF (Figure 3A). Previously, we used this EMSA and indicated that ssDUE-binding activity of ATP–DnaA was specifically stimulated by the DAR (13). Moreover, a pull-down assay revealed that the ATP–DnaA–DAR complex directly binds ssDUE (13).
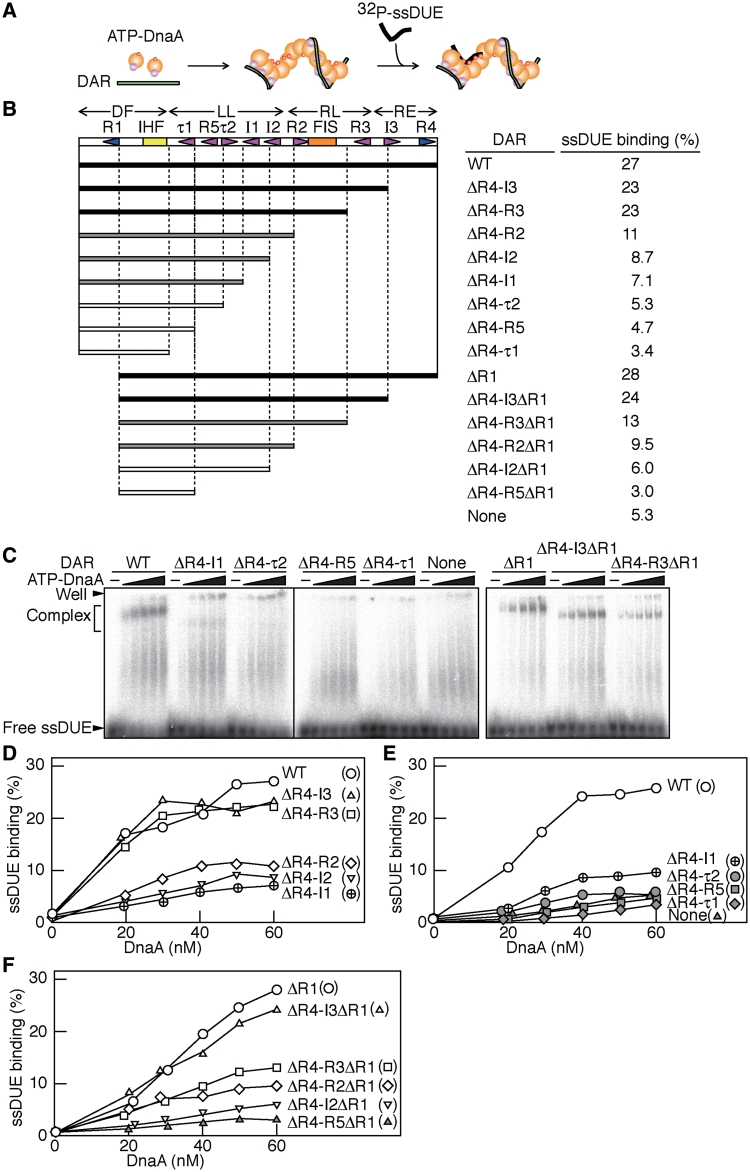
Figure 3.
EMSA for ssDUE-binding activity of ATP–DnaA–DAR complexes. (A) Schematic of the assay. ATP–DnaA was incubated with DAR and end-labeled ssDUE, followed by EMSA. The symbols for DnaA are the same as those used in Figure 2A. 32P-labeled M28 DNA (32P-ssDUE), back bar; DAR derivatives, green bar. An alternative structural model is also conceivable but it is not shown here for the simplicity (see ‘Discussion’ section). (B–F) The motifs within DAR are shown using the same symbols as in Figure 1. (B) ATP–DnaA (0–60 nM) and 32P-ssDUE (2.5 nM) were incubated in the presence or absence (None) of the indicated DAR derivatives (5 nM), followed by EMSA. Representative gel images are shown (C), where – and free ssDUE indicate no DnaA and protein-free ssDUE, respectively. The amounts of ssDUE–ATP–DnaA–DAR complexes (Complex) were quantified using the data shown in (C) and Supplementary Figure S3 and the relative amounts of the complexes to the input ssDUE were plotted as ssDUE binding (%) (D–F). The values obtained using 60 nM ATP–DnaA are shown in (B) (‘ssDUE binding’). Accordingly, the relative activity levels are highlighted using differently grayed bars to indicate the DAR derivatives (closed, 100–80% of the wild-type level; shaded, 50–30% of the wild-type level; open, <25% of the wild-type level) (B).
When the ssDUE containing the 13-mer M and R motifs was incubated in the presence of both wild-type DAR and ATP–DnaA at levels comparable to those used for in vitro reconstituted mini-chromosome replication, 20–30% of the input ssDUE bound to the ATP–DnaA–DAR complexes, forming discrete bands (WT; Figure 3B–E and Supplementary Figure S3). By contrast, in the absence of the DAR, no discrete ssDUE–DnaA complexes were detected (None; Figure 3C and E). We also noticed that, in the presence of DnaA, a small amount of the ssDUE (~7% of the input amount) remained within the gel wells (WT and None; Figure 3C). This could be due to the formation of irregular DnaA aggregates, which then bound to the ssDUE. These aggregates would not be stable, showing smear bands by slow dissociation during electrophoresis. These are consistent with our previous data (13).
Deletions within the right half of the DAR, including the DnaA boxes R3, I3 and R4, maintained ssDUE-binding activity at levels similar to that seen with wild-type DAR (ΔR4-I3 and ΔR4-R3; Figure 3B–E). However, deletion of the DnaA-binding sites R2, I2 and I1 moderately decreased ssDUE-binding activity (ΔR4-R2, ΔR4-I2 and ΔR4-I1; Figure 3B–E). Further deletions of the τ2, R5 and τ1 sites resulted in a further decrease in the activity (ΔR4-τ2, ΔR4-R5 and ΔR4-τ1; Figure 3B–E). These observations are consistent with the idea that the LL region within the DAR plays a crucial role in the formation of ATP–DnaA multimers competent for ssDUE binding.
These data are overall consistent with the data of P1 nuclease assays using the derivatives of pBSoriC and the ori-EcoRI DNA (Figures 1 and 2). Only a slight difference was seen in the results of DnaA box R2 deletion. Whereas DnaA box R2 was dispensable in ssDUE binding of ori-EcoRI derivatives (EC3–ΔR4–ΔR2; Figure 2), this box facilitated the formation of ssDUE binding of DnaA multimers in the EMSA (ΔR4–ΔR2; Figure 3). This difference can be explained by stabilities of DnaA multimers and the difference of the assays (i.e. P1 nuclease assay versus EMSA) in the time taken for yielding the reaction products or the data. DnaA binding to box R2 would become more important for increasing stability of the DnaA multimers formed on the LL region in a co-operative manner, thereby inhibiting the dissociation of these multimers during electrophoresis. This idea is consistent with the fact that DnaA box R2 has a moderate affinity for DnaA (Kd would be 20–40 nM) (9,27) (see ‘Discussion’ section).
We next asked whether DnaA box R1 is required for the formation of ssDUE-binding-competent multimers (Figure 3B, C and F). Notably, the mutant DAR lacking DnaA box R1 (ΔR1) maintained ssDUE-binding activity at the wild-type level (Figure 3B, C and F), indicating that ssDUE-binding DnaA sub-complexes do not include DnaA box R1. Taken together with the data described above and the requirement for DnaA box R1 for ssDUE digestion in the linear oriC-P1 nuclease assay (Figure 2E), we infer that the DnaA sub-complex formed on the LL region is competent in ssDUE binding and the DnaA–DF (box R1) sub-complex recruits ssDUE to the DnaA–LL sub-complex via DNA bending by IHF and interaction between the LL-DF DnaA complexes (see later). This idea can explain that DnaA box R1 plays a specific and crucial role in ssDUE binding of ATP–DnaA–DAR complexes only in cases where DnaA box R1 is in cis to the DUE.
Role for DnaA-bound box R1 and box R3 in DnaA multimerization
The R4 and I3 sites within the RE region were deleted with the box R1 without significant loss of activity (ΔR4–I3ΔR1; Figure 3B and F). Additional deletions, including that of DnaA box R3, moderately inhibited ssDUE- binding activity (ΔR4–R3ΔR1; Figure 3B and F), suggesting that DnaA box R3 within the RL region stimulates the formation of ssDUE-binding-competent ATP–DnaA multimers in the absence of DnaA box R1.
As suggested above, DnaA boxes R2 and R3 would increase the stability of ATP–DnaA multimers formed on the LL region in a co-operative manner. This is consistent with the fact that DnaA box R2 has a moderate affinity for DnaA (Kd would be 20–40 nM) (9,27). The effect of R3-deletion was seen specifically in cases where the DAR lacked DnaA box R1, suggesting that stability of ATP–DnaA multimers formed by binding to DnaA box R2 and its flanking region can be facilitated by either DnaA box R1 or R3 and co-operative binding of DnaA.
It should be also noted that in EMSA, we used the DnaA concentrations ranging from 0 to 80 nM. These DnaA concentrations are reasonable because these are also used for the reconstitution of mini-chromosome replication using purified proteins in vitro (25). Therefore, under our experimental conditions, formation of abnormal DnaA multimers on DAR lacking DnaA boxes R1 and R4 due to extremely high DnaA concentrations is unlikely.
Unique role for the DUE-flanking DnaA box R1 in ssDNA recruitment
We next investigated a unique role for the R1 box. In view of the fact that, when the DUE and box R1 are arranged in cis, IHF supports ssDNA binding of DnaA–oriC complexes (Figure 2F), we hypothesized that the ssDUE is recruited to LL-bound DnaA multimers via IHF-mediated DNA bending within the DF region (Figure 2A). In this context, we further hypothesized that DnaA box R1 (DF region)-bound DnaA directly interacts with the ATP–DnaA multimers formed on the LL region, which enhances their accessibility to the flanking ssDUE, thereby facilitating ssDUE binding and open complex formation.
To test this hypothesis, we performed an EMSA using ATP–DnaA, a number of DAR derivatives, and a 5′-tailed ss-ds oriC DNA fragment containing the ssDUE and dsDNA bearing the DnaA box R1 (ssDUE-R1), or a control non-sense sequence (ssDUE-non) (Figure 4A). ssDUE-R1 formed complexes with the R1 box-deleted DAR (ΔR1) in an ATP–DnaA-dependent manner 2.7-fold more efficiently than ssDUE-non (ΔR1; Figure 4B–E and Supplementary Figure S4). ssDUE-non and ssDUE alone showed a comparable activity in ATP–DnaA-dependent DAR binding (data not shown) (13). These results indicate that DnaA box R1 plays a unique role in promoting ssDUE binding to ATP–DnaA multimers when it is adjacent to the DUE. This is consistent with the above hypothesis that the ssDUE is recruited by DnaA bound to the R1 box and the idea that DnaA sub-complexes formed on oriC have different roles.
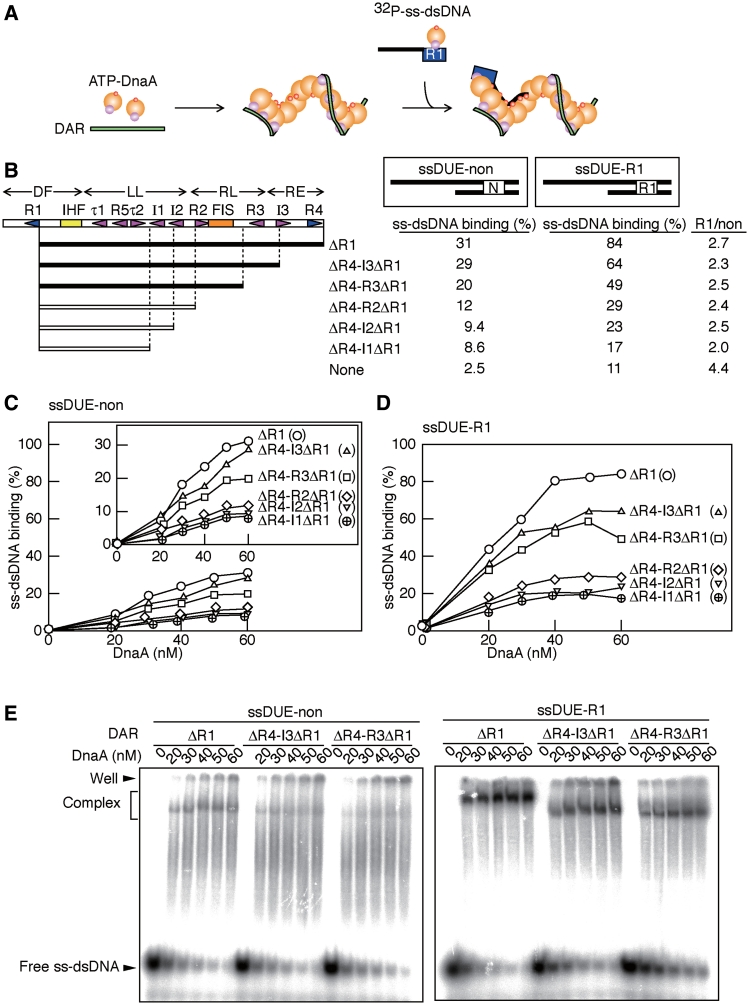
Figure 4.
ssDUE recruitment activity by DnaA box R1 residing in cis to the DUE. (A) A schematic of the assay. Symbols are the same as those used in Figure 3A, except that 32P-labeled ssDUE-R1 and ssDUE conjugated to dsDNA bearing DnaA box R1 (32P-ss-dsDNA) are indicated. An alternative structural model is also conceivable but it is not shown here for the simplicity (see ‘Discussion’ section). (B–E) DAR derivatives, ssDUE-R1 and ssDUE-non are illustrated schematically (R1, R1 box; N, non-sense box) (B). DAR derivatives (5 nM) and ATP–DnaA (0–60 nM) were incubated with 2.5 nM of either 32P-ssDUE-non (C) or ssDUE-R1 (D), followed by EMSA. Representative gel images are shown (E). The amounts of ss-dsDNA–ATP–DnaA–DAR complexes (Complex) were quantified using the data shown in (E) and Supplementary Figure S4 and the relative amounts of the complexes to the input ss-dsDNA were plotted as ss-dsDNA binding (%) (C and D). The values obtained using 60 nM ATP–DnaA are shown in (B) as ss-dsDNA binding. The relative activity levels are highlighted using differently grayed bars to indicate the DAR derivatives (closed, 100–60% of the wild-type level; open, <40% of the wild-type) (B). The ratios of the binding activities of ssDUE-R1 to ssDUE-non (R1/non) are also indicated.
Deletion analysis showed that ΔR4–R3ΔR1 DNA was sufficient to support ssDUE-R1 binding (Figure 4D). This is consistent with the idea that DnaA boxes R3–4 and the I3 site are basically dispensable for the formation of ssDUE-binding-competent ATP–DnaA multimers (Figure 3). Additional deletions, including that of DnaA box R2, decreased ssDUE-R1 binding activity considerably (ΔR4–R2ΔR1; Figure 4D). Also, similar features were seen even in binding of ssDUE-non (Figure 4C). These are consistent with the idea that an ATP–DnaA oligomer, complexed with the LL region, forms a sub-complex that is crucial for ssDUE binding, and is stabilized by DnaA bound to the RL region.
DnaA sub-complexes formed on the DF-LL regions are active in DnaB loading
After unwinding of the duplex DUE, DnaB is loaded onto the resulting ssDUE. This process is mediated by DnaA and DnaC. Previous studies show that binding of multiple DnaAs to oriC is required for a stable interaction with, and loading of, DnaB (7,19,20). Therefore, we investigated whether DnaB loading is dependent on a specific region within the DAR using a form I* assay. In this assay, ssDUE-loaded DnaB helicases further unwind the duplex DNA in the presence of DNA gyrase, resulting in the production of a highly negatively supercoiled form called form I* (39). The results showed that deletion of the RL and RE regions (pBS-ΔR4, pBS-ΔR4–I3 and pBS-ΔR4–R2; Figure 5A and B) maintained form I* formation at ~50% of that seen for the pBS-WT. This suggests that DnaA oligomers, complexed with the DF and LL regions, form a sub-complex associated with baseline activity levels for DnaB loading, and that this activity is stimulated by DnaA oligomers complexed with the RL–RE regions, including the R4 box. This also is consistent with the idea that DnaA sub-complexes formed on oriC have different roles.
Figure 5.
DnaB loading activity of ATP-DnaA complexes formed on the truncated DAR. (A and B) DnaB loading activity was assessed by the form I* assay. The oriC derivatives used in this assay are shown schematically (A). The form I* assay was performed by incubating pBS-WT, or its derivatives (1.6 nM) at 30°C for 15 min in the presence of ATP–DnaA (20 nM) and 0–26 nM of DnaB–DnaC complexes. Gel images and the DNA migration positions of the supercoiled form (I), form I* (I*), the open circular form (II) and linear form (linear) are indicated (B). The relative amounts of form I* and total DNA were quantified and the relative amount of form I* DNA to total DNA was deduced using data obtained using 26 nM DnaBC. The resultant value for the wild-type DNA (pBS-WT) was defined as 1.0 and the relative values for the pBS-WT derivatives are shown in (A) (DnaB loading). Accordingly, the relative activity levels are indicated by grayed bars for the derivatives. (closed, wild-type; shaded, 45–60% of the wild-type level; open, <30% of the wild-type). (C–E) DnaB-binding activity to DnaA–oriC complexes was assessed by a pull-down assay. The biotinylated oriC derivatives used in this pull-down assay are shown schematically (C). ATP–DnaA (1 µM, 10 pmol), HisDnaB (0.5 µM) and DnaC (0.5 µM) were incubated in the presence of the biotinylated oriC derivatives (10 nM), followed by the pull-down assay. Proteins bound to DNA were analyzed by SDS-11% PAGE and silver staining (D). The recovered amounts of proteins (DnaA, DnaB and DnaC) in (D) were determined using standard curves and the values from the negative control (biovector) were subtracted. The recovered amounts of DNA were deduced as described in ‘Materials and Methods’ section. The relative amount of recovered DnaB for the bioWT recovered was defined as 1.0 and those for the bioWT derivatives recovered were deduced and shown as DnaB binding (C). Accordingly, the relative activity levels for the derivatives are indicated by grayed bars as above. Also, the mean numbers and their standard deviations for the recovered protein molecules per the recovered DNA were indicated (E).
Further serial deletions of the I2, I1, τ2, R5 and τ1 sites decreased the formation of form I* to 26%, or less, of that seen for pBS-WT (pBS-ΔR4–I2, pBS-ΔR4–I1, pBS-ΔR4–τ2 and pBS-ΔR4–R5; Figure 5A and B). These results are in broad agreement with those seen in the ori-EcoRI ssDUE digestion assay (Figure 2), rather than with those seen in the supercoiled oriC unwinding assay (Figure 1). pBS-ΔR4-τ2, which was active in DUE unwinding (Figure 1), was virtually inactive in form I* formation. These results suggest that stable ssDUE binding by DnaA oligomers complexed with the LL region is a prerequisite for DnaB loading.
DnaA sub-complexes formed on the RL–RE regions enhance DnaB binding
We also investigated the DnaB-binding activity of the DnaA–DAR sub-complexes by pull-down assay using a biotinylated DNA. In this assay, we used hexahistidine-fused DnaB (HisDnaB) to distinguish DnaA from DnaB by SDS–PAGE. Activities of HisDnaB and intact DnaB were similar in a DnaB-loading assay using mini-chromosome, DnaA, DnaB and DnaC proteins (i.e. the form I* assay), a mini-chromosome replication system reconstituted with purified proteins and an in vivo dnaB mutant-complementation assay (data not shown). ATP–DnaA, HisDnaB and DnaC were co-incubated in the presence of biotinylated DNA bearing the wild-type oriC, its derivative, or a control sequence. The resultant complexes formed on the DNA were isolated using streptavidin beads (Figure 5C and D). Consistent with the previous results (7), HisDnaB and DnaC were assembled with ATP–DnaA multimers on the wild-type oriC DNA (bioWT) (Figure 5C). The number of recovered molecules per bioWT was 9.1 for DnaA, 1.3 for HisDnaB hexamers and 2.0 for DnaC hexamers (Figure 5D), indicating that multiple DnaA molecules on DAR interact with DnaB–DnaC complexes. These numbers are reasonable as an initial complex for replication, although a small number of proteins might be dissociated from the oriC complexes by a washing step in this assay. A deletion of the RL and RE regions (bioΔR4-R2 in Figure 5) sustained DnaA, HisDnaB and DnaC binding at ~50% of that seen for bioWT (Figure 5A, C and D). These indicate that a DnaA–DF–LL sub-complex has a basal affinity for DnaB binding, which is stimulated by DnaA oligomers complexed with the RL–RE regions. This is consistent with the data in the form I* assay showing that DnaA oligomers complexed with the DF–LL region sustain a basal level of DnaB helicase activity (Figure 5A and B).
A further deletion including τ2, and I1–2 sites (bioΔR4-τ2 in Figure 5) also sustained binding of DnaA, DnaB and DnaC at a level similar to that seen for the bioΔR4–R2 (Figure 5A, C and D). Taking into consideration that pBS-ΔR4–τ2 is virtually inactive in the form I* assay (Figure 5A and B) and that τ2, and I1-2 sites within the LL regions are required for ssDUE binding (Figure 2), these results support the idea that stable ssDUE binding, in addition to DnaB binding, by DnaA oligomers complexed with the LL region is crucial for DnaB loading on ssDUE and activation of its helicase function. In these pull-down assays, the recovery of DnaA predominantly depended on oriC DNA, and only a slight amount of DnaA was recovered using a control DNA (biovector), which is most likely due to the nonspecific, weak DNA binding of DnaA and did not cause significant recovery of HisDnaB and DnaC (Figure 5C).
Evolutional conservation of oriC sub-structures for the DUE recruitment
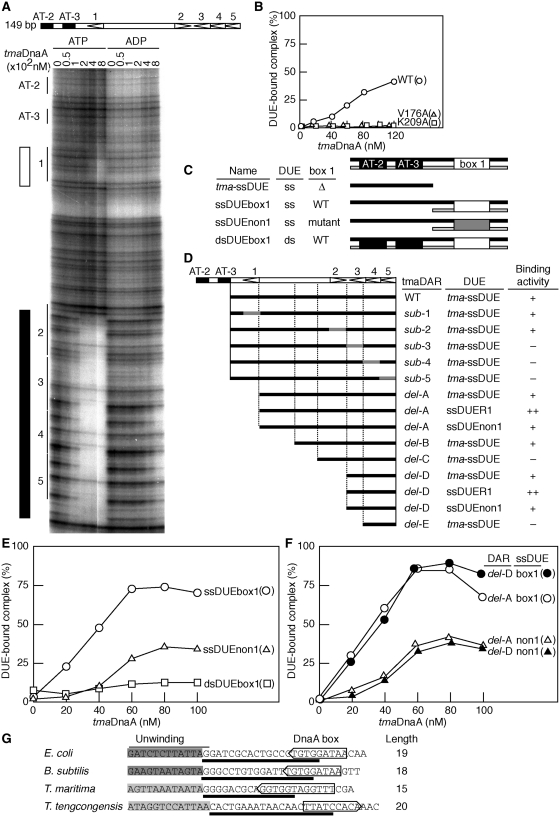
To investigate the evolutionary conservation of the mechanism underlying ssDUE recruitment, we analyzed the initial complexes formed by T. maritima oriC (tma-oriC) and tmaDnaA. A previous study indicated that ATP–tmaDnaA forms a highly ordered complex with tma-oriC more efficiently than ADP–tmaDnaA (40). In the present study, we used DNase I footprint analysis to show that ATP–tmaDnaA interacts with tmaDnaA boxes 2–5 within tma-oriC at protein concentrations of ≥50 nM (Figure 6A) and with box 1 at ≥400 nM. By contrast, ADP–tmaDnaA interacted with boxes 1–5 only weakly, even at 800 nM. These results suggest that ATP–tmaDnaA, but not ADP–tmaDnaA, forms a stable complex on boxes 2–5 in a co-operative manner.
Figure 6.
Specific roles for DnaA boxes in T. maritima oriC. (A) DNase I footprint. ATP– or ADP–tmaDnaA (0–800 nM) and 32P-labeled tma-oriC fragments (10 nM) were incubated with DNase I, followed by sequencing gel analysis. Strongly or weakly protected sites by ATP–tmaDnaA are indicated by closed or open rectangles, respectively. The positions of the DUE (AT-2 and AT-3) and tmaDnaA boxes 1–5 are indicated. (B) EMSA using tmaDnaA, tmaDAR and tma-ssDUE. Various amounts of the wild-type ATP–tmaDnaA (WT) or ATP-forms of tmaDnaA V176A or K209A were incubated with 10 nM of DUE-deleted tma-oriC (tmaDAR WT) DNA, followed by incubation with 2 nM of 32P -labeled tma-ssDUE and electrophoresis on 4% polyacrylamide gels. The relative amounts in the DUE derivatives bound to the tmaDnaA–tmaDAR complexes to those input were deduced as described in Figure 3 and plotted as ‘DUE-bound complex (%)’. For tmaDAR WT, see below. (C) The structures of the DUE derivatives used for EMSA. The DUE derivatives used carry the ssDUE or dsDUE with, or without (Δ), tmaDnaA box1 (WT) or a non-sense box (mutant). Upper-strand, black bar; lower strand, light gray bar; tmaDnaA box 1, open box; non-sense box, shaded box; DUE AT-2 and 3, closed box. (D–F) EMSA using tmaDnaA and derivatives of tmaDAR and tmaDUE. Black and shaded bars indicate DAR wild-type sequences and base substitutions, respectively (D). The DUE derivatives shown in (C) were analyzed as above in the presence of various amounts of wild-type ATP– tmaDnaA and tmaDAR-delA (E) or its derivatives (F). The relative activities for DUE–tmaDnaA–DAR complex formation were deduced as above and plotted. The relative activities (++, >50%; +, 20–50%; −, <20%) for DUE–tmaDnaA–DAR complex formation at 80 nM ATP–tmaDnaA are indicated from experiments using the indicated DAR and DUE derivatives (D). See Supplementary Figure S5 for details. (G) Sequence comparison of an oriC region carrying the DUE and the flanking DnaA box. The arrowed boxes indicate the position and the orientation of the cognate DnaA box flanking the DUE. The unwinding motifs determined in vitro using potassium permanganate modification (dark gray) or P1 nuclease digestion (light gray) are indicated (40–42). Bold lines indicate the number of nucleotides (Length) from the center of the DnaA box to the unwinding motif.
Using a DUE-deleted tma-oriC fragment and EMSA, we also found that ATP-tmaDnaA multimers complexed with tma-oriC specifically bound tma-ssDUE (Figure 6B and C and Supplementary Figure S5) in a similar manner to that of E. coli DnaA and oriC. Moreover, using alanine-substitution mutants of ATP–tmaDnaA, we showed that ssDUE binding requires Val176 within the tmaDnaA H-motif and Lys209 within the B-motif (which correspond to DnaA Val211 and Arg245, respectively, in E. coli) (Figure 6C). These results are in agreement with the findings showing that these mutant tmaDnaAs are inactive in oriC unwinding (13) and that E. coli DnaA V211A and R245A are inactive in ssDUE binding (Figure 2). Taken together, these results are consistent with the idea that tmaDnaA functions in a similar manner to E. coli DnaA during the ATP-dependent conformational activation of DnaA–oriC complexes.
Next, we asked whether a specific region of the tma-oriC DAR is required for ssDUE binding. Mutations within boxes 3–5 inhibited tma-ssDUE binding (sub3-5; Figure 6D and Supplementary Figure S5), whereas mutations in boxes 1 or 2 did not (sub1-2; Figure 6D and Supplementary Figure S5). Consistent with this, deletion analysis showed that the minimal DNA fragment required for tma-ssDUE binding includes boxes 3–5 (del-D; Figure 6 and Supplementary Figure S5), indicating that these boxes are crucial for the formation of an ATP-tmaDnaA oligomer competent for ssDUE-binding.
Finally, we used a 5′-tailed ss-ds tma-oriC fragment containing the ssDUE and dsDNA with box 1 (ssDUEbox1), or a control non-sense sequence (ssDUEnon1), to analyze whether box 1-bound tmaDnaA promoted ssDUE binding of tmaDnaA oligomers formed on boxes 2–5 (Figure 6B, E and F and Supplementary Figure S5). When ATP–tmaDnaA and tma-oriC del-A DNA, including boxes 2–5 but not box 1, were incubated with ssDUEnon1, the ATP–tmaDnaA–tma-oriC del-A complexes bound 20–30% of the input ssDUEnon1 DNA (Figure 6E), a level similar to that seen for tma-ssDUE (Figure 6C). When ssDUEbox1 was used, binding increased 2–3-fold (Figure 6E), suggesting that the DUE-flanking, box 1-bound tmaDnaA promotes the interaction between the ssDUE and the ATP–DnaA multimers formed on boxes 2–5; dynamics similar to those seen for E. coli initial complexes (Figure 4). These observations are consistent with an idea that formation of functionally distinct DnaA sub-complexes on oriC is likely conserved during eubacterial evolution.
We also found that dsDUEbox1 DNA, containing dsDUE and the flanking box 1, was inactive in binding to ATP–tmaDnaA multimers on boxes 2–5 (Figure 6C and E). This suggests that both ssDUE and box 1-bound tmaDnaA are required for binding to ATP–tmaDnaA multimers on boxes 2–5. Efficient binding of ssDUEbox1 to ATP–DnaA was also seen in the presence of tma-oriC del-D (Figure 6F), which is consistent with data showing that box 2 is dispensable for tma-ssDUE binding (Figure 6D). These results support the idea that the DUE-flanking DnaA box shares a common and crucial role in promoting ssDUE recruitment in the E. coli and T. maritima initiation complexes.
It should be noted that, in Thermotoga maritima, Bacillus subtilis and Thermoanaerobacter tengcongensis, whose DNA unwinding regions have been experimentally determined (40–42), the spacing between the unwinding region and the flanking DnaA box is 17 ± 3 bp (bold lines in Figure 6G). This feature is also conserved to that in E. coli (11,41). Also, comprehensive bioinformatic analysis of eubacterial oriC sequences indicates a similar feature (Christoph Weigel, personal communications of unpublished data). These are consistent with the idea that the distance between the DUE and the flanking DnaA box is important for initiation of replication. Indeed, deletions or insertions between the DUE R motif and DnaA box R1 inhibit the ssDUE unwinding in vitro (Figure 2F) and replication initiation in vivo (38). These observations are consistent with the idea that the ssDUE recruitment and the role for DUE-flanking DnaA box in this mechanism are evolutionarily conserved.
DISCUSSION
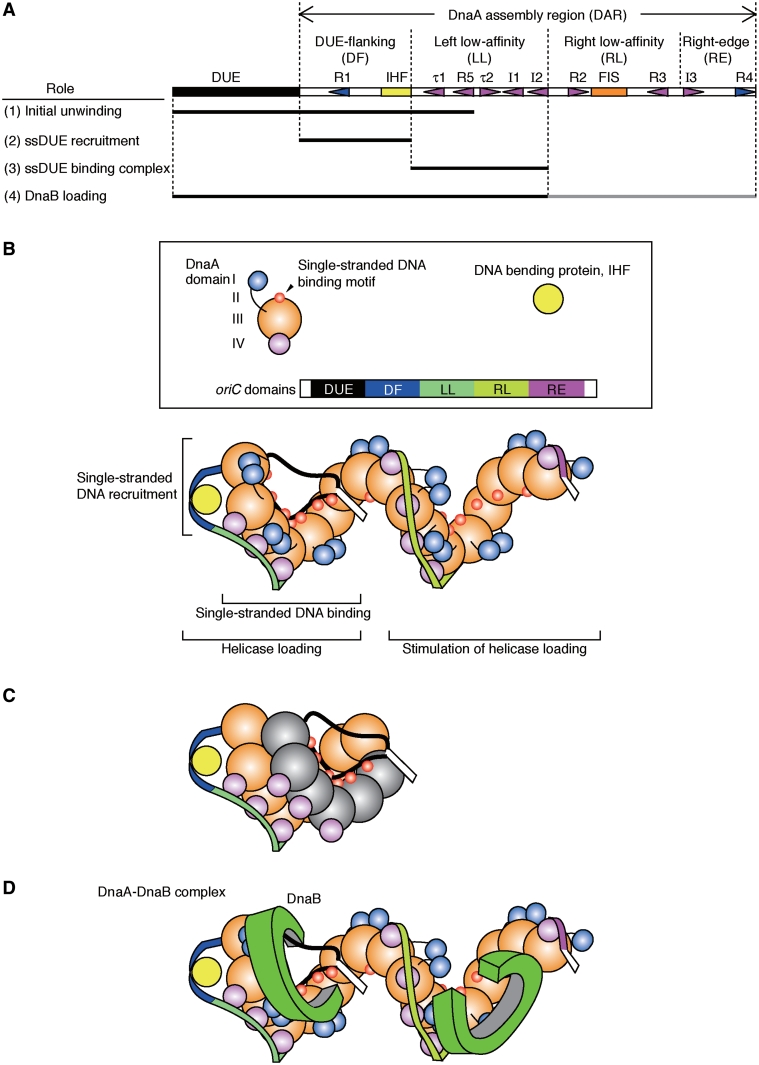
In this study, we report that the DnaA–oriC complex contains functionally distinct sub-complexes that play crucial roles during replicational initiation. ATP–DnaA oligomers complexed with the DAR–DF and –LL regions form an open complex ready for functional DnaB loading. ATP–DnaA oligomers complexed with the LL region are competent for ssDUE binding, which is facilitated by the DnaA–DF DNA sub-complex. This process, termed ssDNA recruitment in this study, is supported by the direct interaction between ATP–DnaA molecules complexed with the DF and LL regions. In other words, the DnaA–DF sub-complex can recruit the unwound ssDUE onto the DnaA–LL sub-complex (Figure 7A and B), thereby promoting the formation of an open complex. Moreover, the resultant complex is a minimum structure competent to DnaB helicase loading onto ssDUE. These findings reveal novel mechanisms and conformational dynamics involved in the formation of initial complexes that have not been previously described.
Figure 7.
Model for oriC unwinding, ssDUE recruitment and helicase loading. (A) Summary figure. Black and gray bars indicate sites bearing predominant and supportive roles, respectively. (B) Model for open complex. The DnaA domains (I, III and IV) and oriC domains (DUE, DF, LL, RL and RE) are distinguished using different colors. The DnaA multimer formed on DAR binds ssDUE. SsDUE is recruited by IHF-dependent DNA bending and interaction of DnaA molecules bound to the DF and LL regions. See text for details. (C) Alternative model for open complex. The DnaA multimer formed on ssDUE interacts with the DnaA multimer formed on DAR. The DnaA domain III within the DnaA–ssDUE complex are colored in gray. Only the DUE-LL regions are shown and the DnaA domains I and II are omitted for the simplicity. (D) Model for helicase loading. DnaA domain I which has a crucial binding site to DnaB is connected to domain II which is a flexible linker to domain III, the AAA+ domain (19). These structural features would support flexible interaction modes and conformational change in oriC–DnaA–DnaB–DnaC complexes. DnaC is omitted in this figure for the simplicity. Similar mechanisms would be possible by the alternative model (C). See text and Supplementary Figure S6 for details.
As mentioned above, the ATP–DnaA sub-complexes formed on the DAR–LL regions are revealed to be a key structure in initial complexes (Figures 2 and 7 A). The formation of the ATP–DnaA–LL sub-complex is facilitated by either (or both) of the adjacent DF and/or RL regions (Figures 3 and 4), likely in a co-operative manner, which is consistent with previous studies (25,43). The DUE and the DAR–DF and –LL regions are fully active during ATP–DnaA-dependent oriC unwinding and ssDUE binding (Figures 1 and 2), indicating that the DF region effectively assists ATP–DnaA–LL sub-complex formation.
We also suggest that DNA bending within the DF region is another key feature in initial complexes (Figure 7A and B). The DF region, containing an intact DnaA box R1 and the IHF-binding site, is required for ssDUE recruitment (Figures 2 and 7 A) suggesting a mechanism involving IHF-induced DNA bending within the DF region, which places R1-bound DnaA and its flanking ssDUE in close proximity to the DnaA–LL sub-complex (Figure 7B). This model is in agreement with reports indicating that R1-bound DnaA and the IHF promote ATP-DnaA assembly on DnaA box R5 and the I1-2 sites within the LL region (32,43). Moreover, this model can give reasonable explanation to the cause of the strict requirement for the spacing between DUE and DnaA box R1 in DUE unwinding (Figure 2F) and in replicational initiation (38).
Our current results do not necessarily exclude a previously proposed model that an unwound complex consists of a continuous DnaA–ssDUE filament adjacent to the DnaA–DAR filament (14,44). This model is supported by a recent report using glutaraldehyde cross-linking experiments which showed that DnaA box-free DnaA molecules can form oligomers on ssDUE (44). However, this ssDUE–DnaA interaction is considerably weak, as indicated in both our previous paper and the present study (Figure 3) (13). DnaA oligomers formed on the DAR–LL region evidently have higher affinity for the ssDUE than DnaA box-free DnaAs; decrease in the amount of free-ssDUE predominantly depends on the DAR (Figure 3C and E; Supplementary Figure S3E). DnaA oligomerization on the DAR would drastically increase its affinity for ssDUE, most likely by creating the regular arrangement of the B/H-motifs and conformational changes within DnaA domain III in an ATP–DnaA–oligomer spiral (6,13). Notably, DnaA oligomers may induce the so-called ‘linkage effect’, which can markedly increase their affinity for the ssDUE by acquiring multiple binding sites (6,8,19,45–47). In addition, whereas several results including those of ssDUE–DnaA–DAR interaction modes could be consistent with the continuous DnaA filament model (Figures 2–4), the strict requirements for IHF binding within DAR and for the spacing between DUE and its flanking DnaA box would be explained quite simply by the ssDUE recruitment model rather than the continuous DnaA filament model. IHF binding within DAR should introduce a break in a DnaA filament, preventing the formation of a DnaA filament continuous from ssDUE to DAR. It would be difficult for the continuous DnaA filament model to give likely explanation for the strict requirement of the spacing, but not specific sequence, between DUE and DnaA box R1. Also, the data of the DNase I footprint experiments of E. coli and T. maritma oriCs suggest that DnaA molecules do not bind to the spacing between DUE and its flanking DnaA box at a significant affinity (Figure 6) (7,8,13,25). Besides, if DnaA forms a multimer on ssDUE and it joins to the DnaA multimer on DAR, the number of DnaA molecules complexed with oriC should increase at open complex formation. However, at least in our experiments, increase in oriC-bound DnaA was not detected during open complex formation (data not shown). Thus, we suggest that ssDUE recruitment to the DnaA multimer on DAR is a plausible mechanism for initiation (Figure 7B). An alternative model that incorporates the possibility of the DnaA oligomerization on ssDUE and meets the strict requirements for IHF and the DUE-to-R1 spacing might be the structure that the DnaA oligomer formed on ssDUE interacts with the DnaA multimer formed on DAR (Figure 7C).
Our study also showed that a truncated oriC carrying only DUE and the DAR–DF and –LL regions can be active in DnaB loading (Figures 5 and 7 A). Moreover, the RL and RE regions were required for full activity in DnaB helicase function. Given that an ATP–DnaA oligomer complexed with the DAR–DF and –LL regions is fully active in oriC unwinding and ssDUE binding (Figures 1 and 2), our results indicate that a DnaA sub-complex formed on the ssDUE-DF-LL regions maintains the basal level of activity for DnaB loading, which is specifically stimulated by a DnaA sub-complex formed on the RL and RE regions (Figure 7A). As the affinity of DnaA monomers for DnaB is faint (48), we suggest that the DnaA oligomer formed on the DAR increases the overall affinity of the DnaA–oriC complexes for DnaB via the ‘linkage effect’ (45–47). Namely, if a single DnaA protomer of a DnaA–oriC complex initially binds a protomer of DnaB hexamer even at a weak affinity, it should impede diffusion of DnaB, stimulating binding of a second DnaA protomer in the complex to another DnaB protomer, which results in marked stimulation in cooperative binding of the proteins and stabilization of the resultant DnaA–DnaB–oriC complexes. In addition, as two DnaB helicases need to be loaded to construct a pair of replication forks at oriC (49), each of the DnaA sub-complexes formed on the DF–LL and RL–RE regions may interact with a single DnaB helicase (Figure 7D; Supplementary Figure S6), thereby promoting the loading of a pair of DnaB helicases and subsequent bidirectional translocation.
Even in vivo, only the left-half of the chromosomal oriC, including the DUE and the DF and LL regions, is essential for initiating chromosomal replication and both the RL and RE regions assist in the initiation at a specific time during the cell cycle (33,50). This indicates that our proposed mechanisms in vitro are basically consistent with in vivo analysis. In the context of our model (Figure 7D; Supplementary Figure S6), in cells lacking the right-half of the chromosomal oriC, a single DnaB helicase can be loaded by DnaA complexes formed on the left-half oriC, expanding ssDNA region unidirectionally. This should allow the second DnaB helicase to load on the generated ssDNA region in a manner similar to the reconstruction of the stalled replication forks (51), thereby establishing bidirectional replication.
Previous experiments using replication cycle-synchronized cultures indicate that the ATP–DnaA level increases to 70–80%, but not 100%, prior to the replicational initiation (52), which suggests a possibility that a mixed complex of ATP– and ADP–DnaA molecules is formed on oriC in vivo to initiate replication. This is consistent with previous reports indicating that only a subgroup of DnaA protomers must be the ATP form for formation of active initiation complexes in vitro (15,25). Based on the present study, we propose that DnaA protomers binding to the RL and RE regions would be compatible with ADP–DnaA without inhibiting the initiation activity (Figures 2–4).
Roles for DnaC in DnaB helicase loading also are consistent with our model. In E. coli, DnaC stably binds DnaB, but not DnaA (7,51). As mentioned above, DnaB hexamer stably binds to the DnaA multimer formed on oriC and thus oriC-DnaA–DnaB–DnaC complexes are formed with a considerable stability (Figure 5) (7). We also revealed using a pull-down assay that like the DnaA multimer on oriC, that on the DAR lacking DUE form stable complexes with DnaB and DnaC (data not shown). As DnaB helicase has a ring-like configuration and a hyperthermophile DnaC-ortholog homooligomer takes on a spiral configuration, it would be possible that binding of the DnaC spiral induces conformational change of the DnaB ring, resulting in opening of the DnaB ring for loading on ssDNA. This assumption is based on an analogy of the opening of the replicase clamp by interaction with the clamp loader complex (18). DnaC and the subunits of the clamp loader complex are members of the AAA+ protein family. The DnaB ring might be inserted in the groove of the DnaA spiral via interaction with multiple molecules of DnaA domain I which carries a specific binding site to DnaB (Supplementary Figure S6) (19). As such, our model is consistent with a possible mechanism of DnaC-dependent DnaB loading. It should be noted that unlike E. coli DnaC, DnaC ortholog from hyperthermophile A. aeolicus can directly interact with the cognate DnaA ortholog (53). The resultant A. aeolicus DnaC–DnaA complex is proposed to promote the cognate DnaB loading on the ssDNA via direct interaction of DnaB with DnaC but not with DnaA. Thus, in A. aeolicus, stable binding between DnaB and the DnaA multimer is not assumed in the process of the DnaB loading, unlike in E. coli (53). This distinct model implies that the role of DnaC in DnaB loading is differentiated during eubacterial evolution. This idea is also consistent with the fact that several eubacterial organisms (e.g. T. maritima) do not contain the cognate DnaC (54).
In E. coli, the high-affinity DnaA boxes R1 and R4 are likely occupied by DnaA throughout the cell cycle (55–57). DnaA assembly on the LL and RL regions is promoted in vivo, possibly due to cell cycle-coordinated increases in the cellular level of ATP–DnaA molecules (52,58). Thus, we suggest that formation of ssDUE-binding-competent ATP–DnaA sub-complexes is an important rate-limiting reaction for the formation of open complexes in vivo.
DiaA protein stimulates the replicational initiation, which is required for the timely initiation during cell cycle (7,8). This protein forms homotetramers which directly binds multiple DnaA molecules and promotes formation of ATP–DnaA–oriC complexes and DUE unwinding in vitro (8). We are currently investigating effects of DiaA in ssDUE recruitment.
As mentioned above, DUE unwinding in DnaA–oriC complexes is effectively stimulated by IHF which causes DNA bending, whereas cells with a scrambled IHF-binding site are viable (33). This is consistent with the facts that E. coli has HU protein, another nucleoid-associated protein causing sharp DNA bending (59), and that HU binds DNA sequence-non-specifically and can stimulate DUE unwinding in vitro of the E. coli and T. maritima oriC plasmids in place of IHF (30,40). Given that HU directly binds to DnaA (60), there is a reasonable possibility that HU might preferentially bind to a site flanking to DnaA box R1 via interaction with DAR-bound DnaA. Orthologs of HU are widely distributed within eubacterial species including T. maritima, whereas some eubacterial species (e.g. T. maritima) do not contain a cognate IHF (29,54).
Like replicational initiation, transcriptional initiation requires the regulated unwinding of duplex DNA and this could be stimulated by DNA bending (61). For instance, the bacterial AAA+ transcriptional stimulators, PspF and NtrC, are suggested to promote DNA bending by binding the σ54-RNA polymerase holoenzyme, thereby stimulating duplex unwinding (62). Therefore, localized DNA-structural changes, such as DNA bending, could be a common mechanism for duplex unwinding in both replication and transcription. Even in eukaryotes, local reorganization of both chromatin and origin DNA is crucial for replication initiation (63,64), which might depend on AAA+ replication proteins.
Like E. coli ATP–DnaA, ATP–tmaDnaA forms oligomers on tma-oriC in a co-operative manner (Figure 6A) and the resultant complex binds the ssDUE, which is stimulated by ssDUE recruitment that is dependent on a DUE-flanking DnaA box (box-1) (Figure 6C–F). Moreover, the B/H-motifs of tmaDnaA are crucial for DUE unwinding and ssDUE binding (Figure 6C) (13). These features are consistent with the idea that the mechanisms underlying open complex formation are evolutionarily conserved among eubacterial initial complexes. Furthermore, we found that ATP–tmaDnaA binding to DnaA box-1 requires higher DnaA concentrations than other DnaA boxes within tma-oriC (Figure 6A). Thus, unlike in E. coli, ssDUE recruitment may be the rate-limiting reaction for open complex formation in T. maritima. Consistent with this, DnaA box-1 is indispensable for DUE unwinding (Ozaki and Katayama, unpublished data). This implies that the order of DnaA assembly might have differentiated during the process of eubacterial evolution, and may be related to the regulatory systems they acquired for adaptation. Similarly, in eukaryotes, the order and timing of replication factors (i.e. GINS, Cdc45 and Polε) associating with the origin have a variety, whereas these factors are highly conserved (65–67).
Besides oriC, E. coli DnaA multimers bind specific genomic loci bearing the clusters of DnaA boxes and excuse various activities for the regulation of initiation (2). These loci include datA for the titration of DnaA, DARS for the reactivation of DnaA, and dnaA promoter for transcriptional auto-regulation (2,9). These observations are consistent with the idea that multiple functions of DnaA are regulated by distinct types of DnaA complexes formed on each DNA region. In other eubacteria, the architecture of oriC is evolutionally differentiated in size and the number of DnaA boxes (9). In many species, the oriC region is longer and bears more DnaA boxes than that in E. coli. These differences could be explained by an idea that functionally different, multiple DnaA sub-complexes are formed on the longer oriC region for DUE unwinding, DnaB loading, DnaA titration and DnaA reactivation.
NOTE ADDED IN PROOF
While this paper was in press, the following paper was published online: Duderstadt,K.E., Chung,K., and Berger,J.M. (2011) DNA stretching by bacterial initiators promotes replication origin opening. Nature, doi:10.1038/nature10455. This paper reports the crystal structure of a truncated DnaA ortholog complexed with ssDNA. The work demonstrates a possible mode of interaction of DnaA oligomers in ssDNA binding. The structure and other data in the paper are fully consistent with the oriC-DnaA complex structure model for the ssDUE recruitment proposed here as well as the continuous DnaA filament model proposed by others previously (14,44). The crystal structure supports the role for B/H-motifs in ssDUE binding.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Materials and Methods, Supplementary Figures S1–6, Supplementary References (7,13,18–19,36,40,68).
FUNDING
Funding for open access charge: Grants-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; and the Japan Society for Promotion of Science.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Christoph Weigel for the sharing of unpublished data, Masahiro Higashi and Kenji Keyamura for assistance in the form I* assay, and the Research Support Center, Graduate School of Medical Sciences, Kyushu University for DNA sequencing.
REFERENCES
- 1.Duderstadt KE, Berger JM. AAA+ ATPases in the initiation of DNA replication. Crit. Rev. Biochem. Mol. Biol. 2008;43:163–187. doi: 10.1080/10409230802058296. [DOI] [PubMed] [Google Scholar]
- 2.Katayama T, Ozaki S, Keyamura K, Fujimitsu K. Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat. Rev. Microbiol. 2010;8:163–170. doi: 10.1038/nrmicro2314. [DOI] [PubMed] [Google Scholar]
- 3.Wigley DB. ORC proteins: marking the start. Curr. Opin. Struct. Biol. 2009;19:72–78. doi: 10.1016/j.sbi.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 4.Kaguni JM. DnaA: controlling the initiation of bacterial DNA replication and more. Annu. Rev. Microbiol. 2006;60:351–375. doi: 10.1146/annurev.micro.60.080805.142111. [DOI] [PubMed] [Google Scholar]
- 5.Leonard AC, Grimwade JE. Regulating DnaA complex assembly: it is time to fill the gaps. Curr. Opin. Microbiol. 2010;13:766–772. doi: 10.1016/j.mib.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ozaki S, Katayama T. DnaA structure, function, and dynamics in the initiation at the chromosomal origin. Plasmid. 2009;62:71–82. doi: 10.1016/j.plasmid.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Keyamura K, Abe Y, Higashi M, Ueda T, Katayama T. DiaA dynamics are coupled with changes in initial origin complexes leading to helicase loading. J. Biol. Chem. 2009;284:25038–25050. doi: 10.1074/jbc.M109.002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keyamura K, Fujikawa N, Ishida T, Ozaki S, Su'etsugu M, Fujimitsu K, Kagawa W, Yokoyama S, Kurumizaka H, Katayama T. The interaction of DiaA and DnaA regulates the replication cycle in E. coli by directly promoting ATP DnaA-specific initiation complexes. Genes Dev. 2007;21:2083–2099. doi: 10.1101/gad.1561207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Messer W. The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication. FEMS Microbiol. Rev. 2002;26:355–374. doi: 10.1111/j.1574-6976.2002.tb00620.x. [DOI] [PubMed] [Google Scholar]
- 10.Baker TA, Kornberg A. Transcriptional activation of initiation of replication from the E. coli chromosomal origin: an RNA–DNA hybrid near oriC. Cell. 1988;55:113–123. doi: 10.1016/0092-8674(88)90014-1. [DOI] [PubMed] [Google Scholar]
- 11.Bramhill D, Kornberg A. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988;52:743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- 12.Sekimizu K, Bramhill D, Kornberg A. ATP activates dnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell. 1987;50:259–265. doi: 10.1016/0092-8674(87)90221-2. [DOI] [PubMed] [Google Scholar]
- 13.Ozaki S, Kawakami H, Nakamura K, Fujikawa N, Kagawa W, Park SY, Yokoyama S, Kurumizaka H, Katayama T. A common mechanism for the ATP-DnaA-dependent formation of open complexes at the replication origin. J. Biol. Chem. 2008;283:8351–8362. doi: 10.1074/jbc.M708684200. [DOI] [PubMed] [Google Scholar]
- 14.Speck C, Messer W. Mechanism of origin unwinding: sequential binding of DnaA to double- and single-stranded DNA. EMBO J. 2001;20:1469–1476. doi: 10.1093/emboj/20.6.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yung BY, Crooke E, Kornberg A. Fate of the DnaA initiator protein in replication at the origin of the Escherichia coli chromosome in vitro. J. Biol. Chem. 1990;265:1282–1285. [PubMed] [Google Scholar]
- 16.Hwang DS, Kornberg A. Opposed actions of regulatory proteins, DnaA and IciA, in opening the replication origin of Escherichia coli. J. Biol. Chem. 1992;267:23087–23091. [PubMed] [Google Scholar]
- 17.Heller RC, Marians KJ. Replisome assembly and the direct restart of stalled replication forks. Nat. Rev. Mol. Cell. Biol. 2006;7:932–943. doi: 10.1038/nrm2058. [DOI] [PubMed] [Google Scholar]
- 18.Indiani C, O'Donnell M. The replication clamp-loading machine at work in the three domains of life. Nat. Rev. Mol. Cell. Biol. 2006;7:751–761. doi: 10.1038/nrm2022. [DOI] [PubMed] [Google Scholar]
- 19.Abe Y, Jo T, Matsuda Y, Matsunaga C, Katayama T, Ueda T. Structure and function of DnaA N-terminal domains: specific sites and mechanisms in inter-DnaA interaction and in DnaB helicase loading on oriC. J. Biol. Chem. 2007;282:17816–17827. doi: 10.1074/jbc.M701841200. [DOI] [PubMed] [Google Scholar]
- 20.Felczak MM, Simmons LA, Kaguni JM. An essential tryptophan of Escherichia coli DnaA protein functions in oligomerization at the E. coli replication origin. J. Biol. Chem. 2005;280:24627–24633. doi: 10.1074/jbc.M503684200. [DOI] [PubMed] [Google Scholar]
- 21.Nozaki S, Ogawa T. Determination of the minimum domain II size of Escherichia coli DnaA protein essential for cell viability. Microbiology. 2008;154:3379–3384. doi: 10.1099/mic.0.2008/019745-0. [DOI] [PubMed] [Google Scholar]
- 22.Erzberger JP, Pirruccello MM, Berger JM. The structure of bacterial DnaA: implications for general mechanisms underlying DNA replication initiation. EMBO J. 2002;21:4763–4773. doi: 10.1093/emboj/cdf496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 24.Fujikawa N, Kurumizaka H, Nureki O, Terada T, Shirouzu M, Katayama T, Yokoyama S. Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res. 2003;31:2077–2086. doi: 10.1093/nar/gkg309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawakami H, Keyamura K, Katayama T. Formation of an ATP-DnaA-specific initiation complex requires DnaA Arginine 285, a conserved motif in the AAA+ protein family. J. Biol. Chem. 2005;280:27420–27430. doi: 10.1074/jbc.M502764200. [DOI] [PubMed] [Google Scholar]
- 26.Erzberger JP, Mott ML, Berger JM. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat. Struct. Mol. Biol. 2006;13:676–683. doi: 10.1038/nsmb1115. [DOI] [PubMed] [Google Scholar]
- 27.Schaper S, Messer W. Interaction of the initiator protein DnaA of Escherichia coli with its DNA target. J. Biol. Chem. 1995;270:17622–17626. doi: 10.1074/jbc.270.29.17622. [DOI] [PubMed] [Google Scholar]
- 28.McGarry KC, Ryan VT, Grimwade JE, Leonard AC. Two discriminatory binding sites in the Escherichia coli replication origin are required for DNA strand opening by initiator DnaA-ATP. Proc. Natl Acad. Sci. USA. 2004;101:2811–2816. doi: 10.1073/pnas.0400340101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dillon SC, Dorman CJ. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 2010;8:185–195. doi: 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- 30.Hwang DS, Kornberg A. Opening of the replication origin of Escherichia coli by DnaA protein with protein HU or IHF. J. Biol. Chem. 1992;267:23083–23086. [PubMed] [Google Scholar]
- 31.Roth A, Urmoneit B, Messer W. Functions of histone-like proteins in the initiation of DNA replication at oriC of Escherichia coli. Biochimie. 1994;76:917–923. doi: 10.1016/0300-9084(94)90016-7. [DOI] [PubMed] [Google Scholar]
- 32.Ryan VT, Grimwade JE, Nievera CJ, Leonard AC. IHF and HU stimulate assembly of pre-replication complexes at Escherichia coli oriC by two different mechanisms. Mol. Microbiol. 2002;46:113–124. doi: 10.1046/j.1365-2958.2002.03129.x. [DOI] [PubMed] [Google Scholar]
- 33.Weigel C, Messer W, Preiss S, Welzeck M, Boye E. The sequence requirements for a functional Escherichia coli replication origin are different for the chromosome and a minichromosome. Mol. Microbiol. 2001;40:498–507. doi: 10.1046/j.1365-2958.2001.02409.x. [DOI] [PubMed] [Google Scholar]
- 34.Wold S, Crooke E, Skarstad K. The Escherichia coli Fis protein prevents initiation of DNA replication from oriC in vitro. Nucleic Acids Res. 1996;24:3527–3532. doi: 10.1093/nar/24.18.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ryan VT, Grimwade JE, Camara JE, Crooke E, Leonard AC. Escherichia coli prereplication complex assembly is regulated by dynamic interplay among Fis, IHF and DnaA. Mol. Microbiol. 2004;51:1347–1359. doi: 10.1046/j.1365-2958.2003.03906.x. [DOI] [PubMed] [Google Scholar]
- 36.Kawakami H, Ozaki S, Suzuki S, Nakamura K, Senriuchi T, Su'etsugu M, Fujimitsu K, Katayama T. The exceptionally tight affinity of DnaA for ATP/ADP requires a unique aspartic acid residue in the AAA+ sensor 1 motif. Mol. Microbiol. 2006;62:1310–1324. doi: 10.1111/j.1365-2958.2006.05450.x. [DOI] [PubMed] [Google Scholar]
- 37.Filutowicz M, Grimek H, Appelt K. Purification of the Escherichia coli integration host factor (IHF) in one chromatographic step. Gene. 1994;147:149–150. doi: 10.1016/0378-1119(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 38.Hsu J, Bramhill D, Thompson CM. Open complex formation by DnaA initiation protein at the Escherichia coli chromosomal origin requires the 13-mers precisely spaced relative to the 9-mers. Mol. Microbiol. 1994;11:903–911. doi: 10.1111/j.1365-2958.1994.tb00369.x. [DOI] [PubMed] [Google Scholar]
- 39.Baker TA, Sekimizu K, Funnell BE, Kornberg A. Extensive unwinding of the plasmid template during staged enzymatic initiation of DNA replication from the origin of the Escherichia coli chromosome. Cell. 1986;45:53–64. doi: 10.1016/0092-8674(86)90537-4. [DOI] [PubMed] [Google Scholar]
- 40.Ozaki S, Fujimitsu K, Kurumizaka H, Katayama T. The DnaA homolog of the hyperthermophilic eubacterium Thermotoga maritima forms an open complex with a minimal 149-bp origin region in an ATP-dependent manner. Genes Cells. 2006;11:425–438. doi: 10.1111/j.1365-2443.2006.00950.x. [DOI] [PubMed] [Google Scholar]
- 41.Krause M, Messer W. DnaA proteins of Escherichia coli and Bacillus subtilis: coordinate actions with single-stranded DNA-binding protein and interspecies inhibition during open complex formation at the replication origins. Gene. 1999;228:123–132. doi: 10.1016/s0378-1119(99)00009-8. [DOI] [PubMed] [Google Scholar]
- 42.Pei H, Liu J, Li J, Guo A, Zhou J, Xiang H. Mechanism for the TtDnaA-Tt-oriC cooperative interaction at high temperature and duplex opening at an unusual AT-rich region in Thermoanaerobacter tengcongensis. Nucleic Acids Res. 2007;35:3087–3099. doi: 10.1093/nar/gkm137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller DT, Grimwade JE, Betteridge T, Rozgaja T, Torgue JJ, Leonard AC. Bacterial origin recognition complexes direct assembly of higher-order DnaA oligomeric structures. Proc. Natl Acad. Sci. USA. 2009;106:18479–18484. doi: 10.1073/pnas.0909472106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duderstadt KE, Mott ML, Crisona NJ, Chuang K, Yang H, Berger JM. Origin remodeling and opening in bacteria rely on distinct assembly states of the DnaA initiator. J. Biol. Chem. 2010;285:28229–28239. doi: 10.1074/jbc.M110.147975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stauffer ME, Chazin WJ. Structural mechanisms of DNA replication, repair, and recombination. J. Biol. Chem. 2004;279:30915–30918. doi: 10.1074/jbc.R400015200. [DOI] [PubMed] [Google Scholar]
- 46.Arunkumar AI, Stauffer ME, Bochkareva E, Bochkarev A, Chazin WJ. Independent and coordinated functions of replication protein A tandem high affinity single-stranded DNA binding domains. J. Biol. Chem. 2003;278:41077–41082. doi: 10.1074/jbc.M305871200. [DOI] [PubMed] [Google Scholar]
- 47.Zhou HX. The affinity-enhancing roles of flexible linkers in two-domain DNA-binding proteins. Biochemistry. 2001;40:15069–15073. doi: 10.1021/bi015795g. [DOI] [PubMed] [Google Scholar]
- 48.Sutton MD, Carr KM, Vicente M, Kaguni JM. Escherichia coli DnaA protein. The N-terminal domain and loading of DnaB helicase at the E. coli chromosomal origin. J. Biol. Chem. 1998;273:34255–34262. doi: 10.1074/jbc.273.51.34255. [DOI] [PubMed] [Google Scholar]
- 49.Fang L, Davey MJ, O'Donnell M. Replisome assembly at oriC, the replication origin of E. coli, reveals an explanation for initiation sites outside an origin. Mol. Cell. 1999;4:541–553. doi: 10.1016/s1097-2765(00)80205-1. [DOI] [PubMed] [Google Scholar]
- 50.Stepankiw N, Kaidow A, Boye E, Bates D. The right half of the Escherichia coli replication origin is not essential for viability, but facilitates multi-forked replication. Mol. Microbiol. 2009;74:467–479. doi: 10.1111/j.1365-2958.2009.06877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schaeffer PM, Headlam MJ, Dixon NE. Protein–protein interactions in the eubacterial replisome. IUBMB Life. 2005;57:5–12. doi: 10.1080/15216540500058956. [DOI] [PubMed] [Google Scholar]
- 52.Kurokawa K, Nishida S, Emoto A, Sekimizu K, Katayama T. Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. EMBO J. 1999;18:6642–6652. doi: 10.1093/emboj/18.23.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mott ML, Erzberger JP, Coons MM, Berger JM. Structural synergy and molecular crosstalk between bacterial helicase loaders and replication initiators. Cell. 2008;135:623–634. doi: 10.1016/j.cell.2008.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nelson KE, Clayton RA, Gill SR, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Nelson WC, Ketchum KA, et al. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 55.Cassler MR, Grimwade JE, Leonard AC. Cell cycle-specific changes in nucleoprotein complexes at a chromosomal replication origin. EMBO J. 1995;14:5833–5841. doi: 10.1002/j.1460-2075.1995.tb00271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nievera C, Torgue JJ, Grimwade JE, Leonard AC. SeqA blocking of DnaA-oriC interactions ensures staged assembly of the E. coli pre-RC. Mol. Cell. 2006;24:581–592. doi: 10.1016/j.molcel.2006.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Samitt CE, Hansen FG, Miller JF, Schaechter M. In vivo studies of DnaA binding to the origin of replication of Escherichia coli. EMBO J. 1989;8:989–993. doi: 10.1002/j.1460-2075.1989.tb03462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fujimitsu K, Senriuchi T, Katayama T. Specific genomic sequences of E. coli promote replicational initiation by directly reactivating ADP-DnaA. Genes Dev. 2009;23:1221–1233. doi: 10.1101/gad.1775809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Swinger KK, Lemberg KM, Zhang Y, Rice PA. Flexible DNA bending in HU-DNA cocrystal structures. EMBO J. 2003;22:3749–3760. doi: 10.1093/emboj/cdg351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chodavarapu S, Felczak MM, Yaniv JR, Kaguni JM. Escherichia coli DnaA interacts with HU in initiation at the E. coli replication origin. Mol. Microbiol. 2008;67:781–792. doi: 10.1111/j.1365-2958.2007.06094.x. [DOI] [PubMed] [Google Scholar]
- 61.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends. Biochem. Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 62.Ghosh T, Bose D, Zhang X. Mechanisms for activating bacterial RNA polymerase. FEMS Microbiol. Rev. 2010;34:611–627. doi: 10.1111/j.1574-6976.2010.00239.x. [DOI] [PubMed] [Google Scholar]
- 63.Hayashi MT, Masukata H. Regulation of DNA replication by chromatin structures: accessibility and recruitment. Chromosoma. 2011;120:39–46. doi: 10.1007/s00412-010-0287-4. [DOI] [PubMed] [Google Scholar]
- 64.Remus D, Diffley JF. Eukaryotic DNA replication control: lock and load, then fire. Curr. Opin. Cell Biol. 2009;21:771–777. doi: 10.1016/j.ceb.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 65.Labib K. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 2010;24:1208–1219. doi: 10.1101/gad.1933010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tanaka S, Araki H. Regulation of the initiation step of DNA replication by cyclin-dependent kinases. Chromosoma. 2010;119:565–574. doi: 10.1007/s00412-010-0291-8. [DOI] [PubMed] [Google Scholar]
- 67.Yabuuchi H, Yamada Y, Uchida T, Sunathvanichkul T, Nakagawa T, Masukata H. Ordered assembly of Sld3, GINS and Cdc45 is distinctly regulated by DDK and CDK for activation of replication origins. EMBO J. 2006;25:4663–4674. doi: 10.1038/sj.emboj.7601347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nash HA, Robertson CA, Flamm E, Weisberg RA, Miller HI. Overproduction of Escherichia coli integration host factor, a protein with nonidentical subunits. J. Bacteriol. 1987;169:4124–4127. doi: 10.1128/jb.169.9.4124-4127.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.