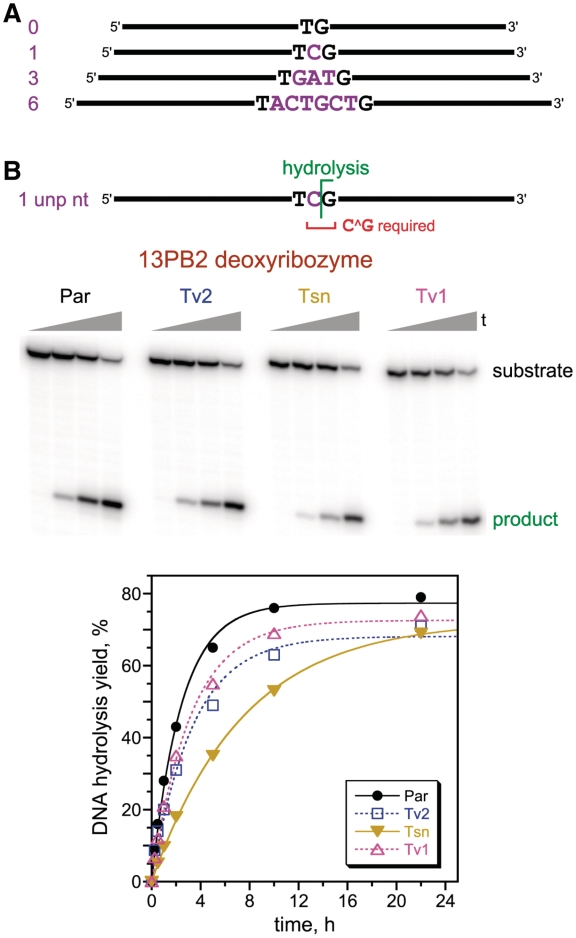
Figure 2.
Selections with DNA substrates that have 0, 1, 3, or 6 unpaired nucleotides between the two duplex binding arms. (A) Substrates, showing the unpaired nucleotides (purple). The complete sequences of the binding arms (black) are provided in the Experimental Section. Compare to Figure 1B for structural context of the substrate. The corresponding selections were designated PA–PD. (B) Single-turnover in trans assays of the 13PB2 deoxyribozyme, which was identified from the selection with 1 unpaired nucleotide. 13PB2 requires only C^G at the cleavage site and forms 3′-hydroxyl + 5′-phosphate termini. On the PAGE image are shown representative time points at t = 30 s, 15 min, 2 h and 22 h. The activity is maintained well when all nucleotides of the DNA substrate outside of the cleavage-site C^G are changed from the parent (Par) sequence via systematic changes: transitions (Tsn: A↔G and T↔C); transversions-1 (Tv1: A↔C and G↔T); transversions-2 (Tv2: A↔T and G↔C). All nucleotides in the deoxyribozyme binding arms (except opposite the unchanged C^G of the substrate) were covaried to maintain Watson-Crick base pairing with the substrate. kobs values (h−1): Par 0.41, Tv2 0.30, Tsn 0.13, Tv1 0.32. The 13PB2 sequence is given in Figure 3. 13PB2 also has moderate activity with G^G rather than C^G substrates (Supplementary Figure S3).