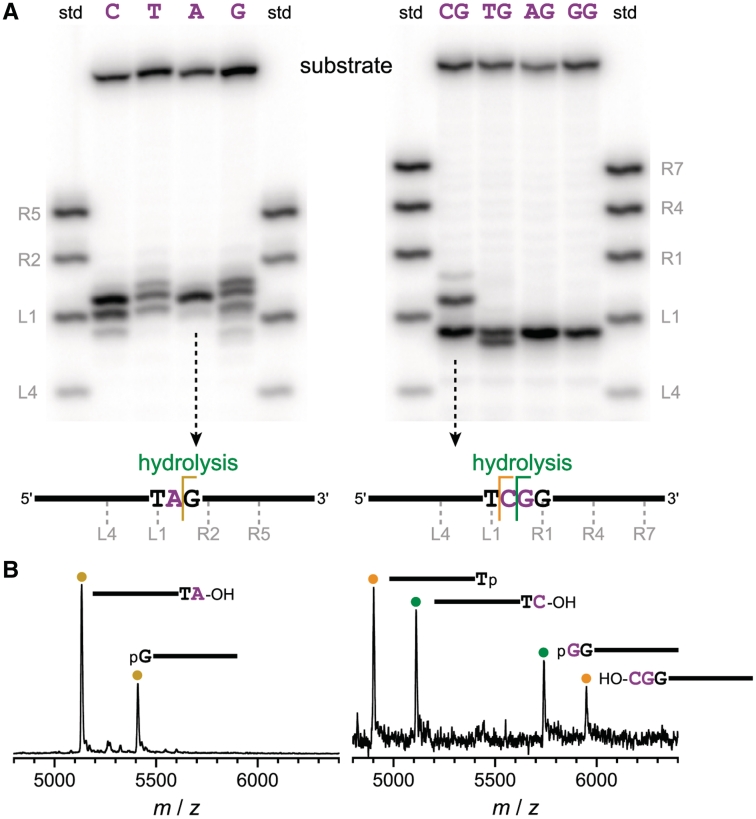
Figure 4.
Outcomes of systematic selections with DNA substrates that have one or two unpaired nucleotides, as revealed by single-turnover in trans assays of uncloned selection pools. For substrates with one unpaired nucleotide X, selections were designated RV–RY; in cis uncloned pool yields for round 7 were 37, 27, 37 and 18%. For substrates with two unpaired nucleotides XG, selections were designated RQ–RT; in cis uncloned pool yields for round 8 were 44, 43, 56 and 28%. (A) PAGE assays using 5′-32P-radiolabeled substrates and standards. The unpaired nucleotide(s) of the substrate are shown above each lane. The 3′-OH standards are each designated with their 3′-terminal nucleotide, where L1 is the first paired nucleotide immediately 5′ of the unpaired nucleotide(s) and R1 is the first paired nucleotide immediately 3′ of the unpaired nucleotide(s). (B) Representative MALDI mass spectrometry data for the two indicated uncloned pools. Each assigned peak has observed mass within 0.05% of the calculated value (data not shown). DNA hydrolysis was predominant in each uncloned pool, although a small amount of deglycosylation-induced cleavage was also evident in some cases. For example, the selection with a single unpaired G gave a mixture of hydrolysis and deglycosylation products, as assessed by mass spectrometry (Supplementary Figure S6).