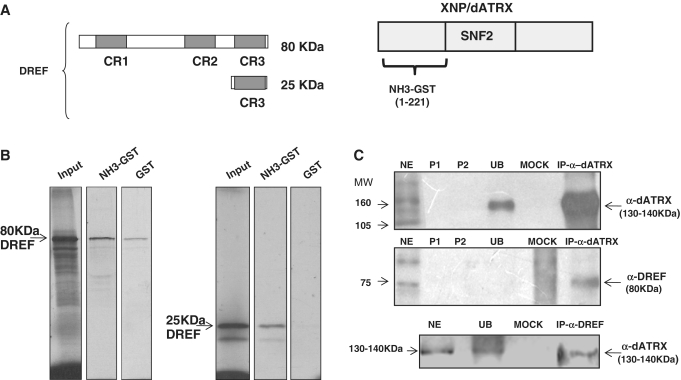
Figure 1.
Physical interaction between XNP/dATRX and DREF. (A) Representation of the fragments assayed by pull down, the three conserved regions of DREF are shown (gray boxes), CR3 corresponds to the region that interacts with XNP/dATRX in the two-hybrid assay. On the right side, the 221 amino acids of XNP/dATRX used for the two-hybrid and the pull-down assays is shown. (B) Pull-down assay. For both panels: the first lane shows the amount of transcription/translation labeled protein (full-length DREF or the CR3 domain) used for each experiment (input), the second lane is the experimental interaction (GST-dATRX and dDREF) and the third lane is the control interaction (GST and dDREF) for each analyzed polypeptide. (C) CoIP of XNP/dATRX and DREF. Transcriptional active nuclear fraction from adult flies or 0–24 h embryos (NE); P1 is the pre-clearing 1; P2 is pre-clearing 2; UB correspond to the unbound protein fraction; Mock is the incubation with an unrelated antibody, the upper and middle panels correspond to an IP performed against XNP/dATRX. Upper panel was blotted with α-dATRX, middle panel with α-dDREF and the lower panel again with α-dATRX after an IP against DREF.