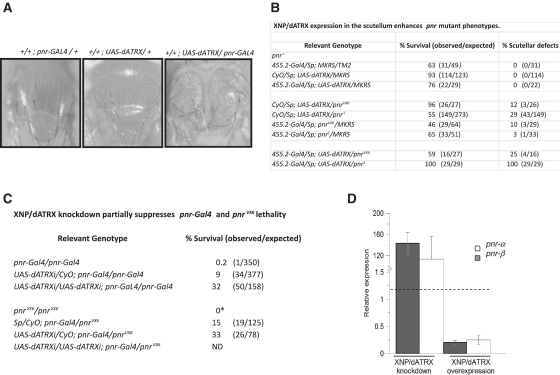
Figure 2.
Modification of pnr mutant phenotypes by XNP/dATRX dosage. (A) Enhancement of the thoracic cleft phenotype by the expression of the UAS-dATRX transgene in the pnr-expression domain. The genotypes of each organism are indicated at the top. Note the enhancement of the thorax cleft defect in the UAS-dATRX/pnr-Gal4 fly. This result is in agreement with Peña-Rangel et al. (39) where they used a different P-UAS construct that overexpresses XNP/dATRX in the pnr expression domain (27). (B) Enhancement of pnrVX6 and pnr1 scutellar defects by the expression of theUAS-dATRX transgene in the scutellum. Relevant genotypes are indicated. The scutellar defects consists of a reduction in the scutellum size and deformations in the scutellar border (data not shown). No defects in the macrochaetes were observed in these flies. (C) XNP/dATRX depletion rescues the lethality of pnr mutant flies. *Homozygous pnrVX6 flies die as embryos (40). Homozygous pnr-Gal4 or pnr-Gal4/pnrVX6 adult flies survive in the presence of one or two copies of the UAS-dATRXi transgene. (D) Modification of pnr transcript levels by XNP/dATRX. Expression of transgenes was driven by the Act5C-Gal4 driver. Depletion of XNP/dATRX by ectopic expression of the UAS-dATRXi transgene increases the transcript levels of the two pnr isoforms in Drosophila embryos. On the contrary, ectopic expression of XNP/dATRX by the UAS-dATRX transgene, reduced bothpnr mRNA levels. The quantification of the mRNA of mutant embryos was performed by RT–qPCR compared with the relative mRNA levels of wild-type embryos of the same age. The relative expression levels are adjusted to one, which is the level of each pnr isoform transcript in wild-type flies. Data are from three different experiments.