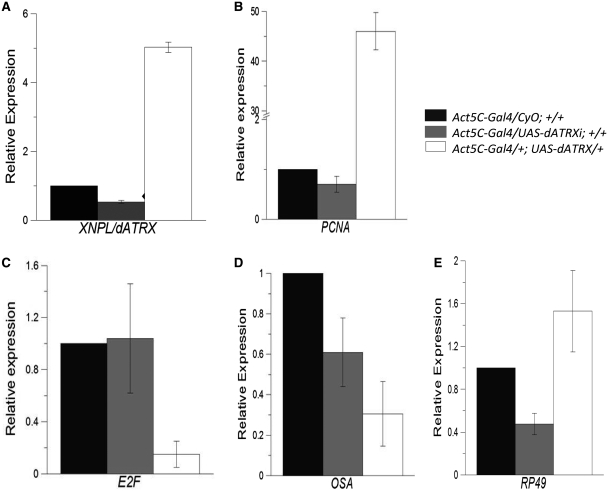
Figure 5.
Effect on the mRNA levels of different genes regulated by DREF by the overexpression and depletion of XNP/dATRX. Total RNA from flies either overexpressing XNP/dATRX or an XNP/dATRX RNAi construct under the control of the Act5C-Gal4 driver (ectopic expression) or without it, was purified from embryos and used in quantitative RT–PCR analysis for different gene transcripts. The genotypes are indicated for each bar color. Black bars indicate the quantification of the specific mRNA in flies with only the Act5C-Gal4driver; gray bars indicate flies expressing the XNP/dATRX RNAi and white bars correspond to flies overexpressing XNP/dATRX. As an internal control we used the mRNA levels of the Dmp8 transcript, which is not affected by the XNP/dATRX levels. The relative expression values are adjusted to 1, which correspond to Act5C-Gal4/CyO; +/+ organisms. (A) Quantification of mRNA levels of XNP/dATRX. Note that the levels of the specific mRNA is reduced when the RNAi against XNP/dATRX is expressed (RNAi-dATRX/Act5C-Gal4) and increased by the overexpression of XNP/dATRX in the UAS-XNP/dATRX (UY3132 line). (B–E) Quantification of the mRNA levels of E2F; osa; PCNA; rp49 in flies overexpressing XNP/dATRX or a RNAi against XNP/dATRX. In all cases the analyzed RNA was prepared from three independent experiments and each cDNA preparation amplified three times for posterior qRT–PCR.