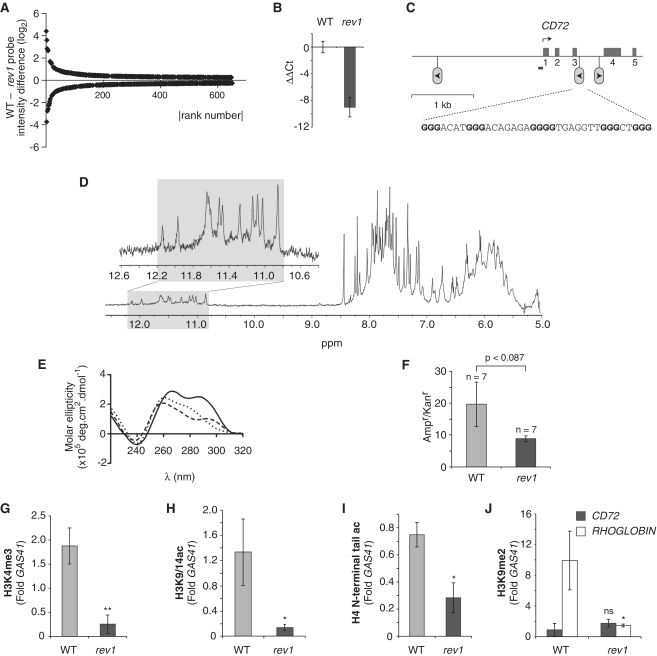
Figure 1.
Epigenetic instability in the CD72 locus of rev1 cells. (A) Plot of probe intensity differences between rev1 and WT ranked by absolute value (for probes whose expression is altered by >0.25 log2 with P < 0.05). (B) qRT-PCR confirming reduction of CD72 expression in a rev1 line independent of that used in the microarray analysis. Error bars show standard deviation. P < 0.05 (2-tailed T-test). (C) Map of the CD72 locus. The first five exons are shown as mid-gray boxes and the positions of the three G4 DNA motifs as inverted lollipops. The arrowhead within the lollipop indicates the direction that a replication fork would have to be travelling for the G-quadruplex structure to form on the leading strand template. The G4 DNA sequence (CD72-F) in the correct orientation to stall replication in rev1 cells is shown, with the dG repeats in bold. The position of the amplicon generated by the ChIP primers (CD72promF & R) is shown as a bar. (D) Part of the 1H NMR spectra of CD72-F. In the inset: an expansion of the spectrum focusing on the imino protons of the guanines involved in a tetrad (gray shaded area). (E) CD spectrum of CD72-F recorded in the presence of LiCl (dotted line), NaCl (dashed line) and KCl (solid line). (F) Replication efficiency of CD72-F incorporated into the leading strand template of the replicating plasmid pQ, shown as the ratio of Ampr to Kanr E. coli colonies. Ampr plasmid contains the CD72-F oligo while Kanr plasmid does not (14). The error bars represent standard error of the mean. P-value calculated with the unpaired t-test. (G) Trimethylation of H3K4 at the CD72 promoter in WT and rev1 cells. (H) Acetylation of H3K9 and K14 at the CD72 promoter in WT and rev1 cells. (I) Acetylation of the H4 N-terminal tail at the CD72 promoter in WT and rev1 cells. (J) Dimethylation of H3K9 at the CD72 and RHOGLOBIN promoters in WT and rev1 cells. Error bars in F–I represent standard error of the mean; P-values compared to WT (one-tailed, unpaired t-test): ns = not significant, *P < 0.075, **P < 0.01.