Abstract
Microsatellite DNA synthesis represents a significant component of human genome replication that must occur faithfully. However, yeast replicative DNA polymerases do not possess high fidelity for microsatellite synthesis. We hypothesized that the structural features of Y-family polymerases that facilitate accurate translesion synthesis may promote accurate microsatellite synthesis. We compared human polymerases κ (Pol κ) and η (Pol η) fidelities to that of replicative human polymerase δ holoenzyme (Pol δ4), using the in vitro HSV-tk assay. Relative polymerase accuracy for insertion/deletion (indel) errors within 2–3 unit repeats internal to the HSV-tk gene concurred with the literature: Pol δ4 >> Pol κ or Pol η. In contrast, relative polymerase accuracy for unit-based indel errors within [GT]10 and [TC]11 microsatellites was: Pol κ ≥ Pol δ4 > Pol η. The magnitude of difference was greatest between Pols κ and δ4 with the [GT] template. Biochemically, Pol κ displayed less synthesis termination within the [GT] allele than did Pol δ4. In dual polymerase reactions, Pol κ competed with either a stalled or moving Pol δ4, thereby reducing termination. Our results challenge the ideology that pol κ is error prone, and suggest that DNA polymerases with complementary biochemical properties can function cooperatively at repetitive sequences.
INTRODUCTION
Repetitive DNA sequences make up at least 50% of the human genome, and many correspond to a DNA structural complexity that is not found in other regions of the genome (1). Microsatellites, one type of interspersed tandem repeat, are ubiquitous throughout the genome (2) and can be important functional regulators of gene expression and phenotypic variability (3). Microsatellite allele length polymorphisms have been implicated as genetic risk factors in human disease (4,5), and expansions at specific microsatellites are responsible for at least 40 inherited human disorders (6). Despite their importance for genome function and disease association, the fidelity mechanisms needed to maintain repetitive sequence stability have not been fully elucidated.
Stability of the eukaryotic genome requires the cooperative activity of multiple DNA polymerases (7,8). The Y-family DNA polymerases are a distinct group of enzymes and are present in bacteria, archaea and eukarya species (9). Initial biochemical characterization of the eukaryotic enzymes focused on their involvement in translesion synthesis (TLS), guided by the homology of eukaryotic genes to Escherichia coli genes that are involved in the DNA damage response. The identification of Pol η as the gene product affected in xeroderma pigmentosum-variant patients (10), together with the biochemical demonstration that Pol η promotes error-free bypass of several types of DNA lesions (11,12), solidified the concept that eukaryotic Y-family polymerases promote genetic stability through accurate TLS. Y-family polymerases can be distinguished by their substrate preferences, in that the enzymes catalyze error-free TLS for a variety of DNA lesions but with differential efficiencies (7). For example, Pol κ has been implicated in the accurate bypass of adducts at the exocyclic N2 position of guanine, such as benzo[a]pyrene diolepoxide (13) and furfuryl (14) lesions. The abilities of Pols κ and η to catalyze DNA synthesis past specific DNA lesions with high fidelity demonstrates that inherent polymerase accuracy is dependent upon the structure of the DNA substrate.
More recently, several investigations have garnered support for more widespread roles of Y-family polymerases in genome function. Human cells deficient in Pol η display aberrant S-phase progression and increased chromosomal fragility in the absence of exogenous exposure to DNA damaging agents (15). Genetic knock-down of Pol η or Pol κ increases the sensitivity of human cells to telomestatin, a compound that stabilizes G tetraplex DNA secondary structures (16). In human fibroblasts, complete DNA synthesis associated with nucleotide excision repair requires both Pol κ and Pol δ as well as Pol ε (17). The spontaneous germline mutation rate within two highly repetitive, expanded short tandem repeat alleles is higher for Pol κ-deficient mice, relative to isogenic Pol κ wild-type mice (18). Although limited, these studies suggest that Y-family polymerases have roles in genome maintenance beyond TLS.
DNA polymerases δ and ε are generally assumed to be responsible for the bulk of DNA replication as well as DNA synthesis associated with repair and recombination (19). These polymerases display highly accurate in vitro DNA synthesis using undamaged DNA templates (20,21). However, we reported that the replicative holoenzyme forms of yeast Pol δ and Pol ε do not possess high fidelity during in vitro microsatellite DNA synthesis (22). Because Y-family polymerases perform accurate TLS of specific lesions, we hypothesized that the structural or biochemical features underlying accurate TLS may also allow increased polymerase discrimination of alternative DNA structures, such as those formed by repetitive microsatellite sequences. In this study, we demonstrate that the human Y-family polymerase, Pol κ, displays a high accuracy during dinucleotide microsatellite DNA synthesis. We postulate that accurately maintaining microsatellites and possibly other repeated DNA sequences may be an additional cellular function for specialized DNA polymerases.
MATERIALS AND METHODS
In vitro gap-filling HSV-tk mutagenesis assay
The 4-subunit recombinant human Pol δ4 and proliferating cell nuclear antigen (PCNA) were purified as described (23). Purified full-length Pol κ (99 kDa) and Pol η (78 kDa) were purchased from Enzymax (Lexington, KY, USA). Microsatellite-containing Herpes simplex virus type 1 thymidine kinase (HSV-tk) vectors have been previously described (24,25). The MluI (position 83) to StuI (position 180) HSV-tk target sequence within the gapped duplex (GD) molecule contains 89–91 bp of HSV-tk gene coding region sequence, and 8 bp ([T]8), 20 bp ([GT]10), or 22 bp ([TC]11) of microsatellite sequence (Figure 1A). In vitro gap-filling reactions (Figure 1B) contained 0.075–0.125 pmol of gapped substrate and 250 µM deoxynucleotide triphosphates (dNTPs) in 100 µl final volume. Pol δ4 reactions contained 40 mM Tris pH 7.8, 10 mM MgCl2, 1 mM DTT, 5 mM NaCl, 200 µg/ml BSA and 1.2–4 pmol pol δ4. Pol δ4 reactions containing accessory factors were supplemented with 2 mM ATP, 40 ng (450 fmol) PCNA and 5 ng (17 fmol) replication factor C (RFC). Reactions were incubated at 37°C for 30 min–1 h. Pol κ and Pol η reactions contained 25 mM potassium phosphate buffer pH 7.2, 5 mM MgCl2, 5 mM DTT, 100 ug/mL BSA, 10% glycerol and 1.9–9.4 pmol of polymerase. Reactions were incubated at 37°C for 2 h. All reactions were terminated with 15 mM Ethylenediaminetetraacetic acid (EDTA), and exchanged into Tris-EDTA (TE) buffer. Complete gap filling by all three polymerases was verified as described (22) (Supplementary Figure S1). An aliquot of DNA from complete gap-filling reactions was used to transform E. coli strain FT334 for mutant frequency determination (26) (Figure 1B). To control for pre-existing mutations, we also determined the HSV-tk mutant frequencies (MF) for each ssDNA used to construct the GD molecules. The DNA sequences within the target region of independent mutants isolated from 2 to 3 polymerase reactions per template were determined (24).
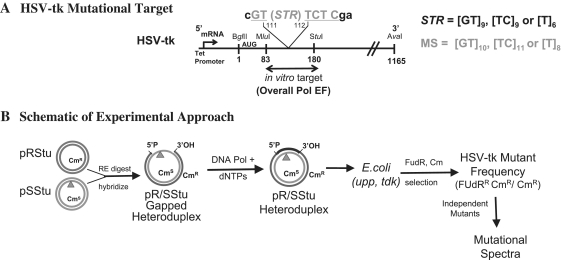
Figure 1.
Schematic of the HSV-tk experimental system. (A) HSV-tk mutational target. Short tandem repeat (STR) sequences were inserted in-frame between bases 111 and 112 of the HSV-tk mutational target to create artificial microsatellites (MS). Inactivating mutations can arise within the MS sequence (MS mutation frequency in Tables 1 and 2), as well as within the approximately 100 base pair region (MluI–StuI) of the in vitro HSV-tk gene mutational target (HSV-tk coding mutation frequency in Tables 1 and 2). The sum of these mutations is shown as the overall Pol EF in Tables 1 and 2. (B) Cartoon of experimental approach. HSV-tk gene cassettes were cloned into two sister plasmids, one of which encodes a functional chloramphenicol acetyltransferase (cat) gene (pRStu) and one which encodes a nonfunctional cat gene (pSStu) (24,25). The location of the MS sequences is indicated by an inverted triangle. Gapped heteroduplex molecules were created by hybridizing the MluI–StuI large fragment from the pRStu vector to ssDNA derived from pSStu vectors. Gel purified gapped substrates were used as templates for DNA synthesis reactions containing purified human DNA polymerases. Product DNAs were introduced into E. coli (upp, tdk) for mutational analyses. Chloramphenicol (Cm) selects for bacteria derived from the heteroduplex CmR strand; FUdR selects for HSV-tk-deficient bacteria. DNA sequence changes of independent FUdRR mutants are determined to derive a mutational spectrum.
Determination of polymerase error frequencies
Pol η and Pol κ produced multiple mutational events per target sequence, while Pol δ4 normally created only 1 mutational event per target. In order to properly compare error rates among these three polymerases, we identified those mutational events that were detectable as single mutational events, and adjusted the HSV-tk MF to reflect multiple errors per target. First, polymerase error frequencies (Pol EFs) were determined by the following equation: Pol EF = (Observed MF)–(ssDNA background MF)–(Outside target MF) where outside target MF is the frequency of errors occurring outside the gap target, either through background errors in the GD molecule, polymerase displacement synthesis or polymerase exonuclease activity (Pol δ4). Next, the Pol EF was adjusted by performing calculations as described (27) with a few modifications. Each mutational event was scored as detectable or undetectable. All frameshifts and those base substitutions that changed an amino acid were considered detectable. Each mutational event was also scored as tandem or non-tandem. Tandem events were those adjacent to one another, whereas non-tandem were errors >1 nt apart. To fully represent all of the mutational events created by the human polymerases, both detectable and undetectable events within the HSV-tk gene are depicted in Supplementary Figure S2. However, only those that were detectable were used for determining Pol EFest. Pol EFs were then corrected for the existence of multiple mutations according to the following formula:
 |
where n is the number of detectable non-tandem errors. The Pol EFest of a specific type of mutational event was calculated from the proportion of the specific mutational event (among the total analyzed) multiplied by Pol EFest.
Pol δ4 and Pol κ primer extension competition analyses
In a common buffer (25 mM potassium phosphate buffer pH 7.2, 5 mM MgCl2, 2.5 mM DTT, 200 µg/ml non-acetylated BSA and 250 µM dNTPs), Pol κ is 4- to 5-fold more active than Pol δ4 in the absence or presence of PCNA, using circular ssDNA as the template (Supplementary Figure S3, panel B). Primer extension analyses were performed to determine termination within a [GT]10 microsatellite for Pol δ4 and Pol κ individually and together in a competition assay. Primer IHTW02, a 15-mer oligonucleotide that initiates synthesis at HSV-tk position 132 was radioactively 5′ end-labeled and hybridized to [GT]10-containing ssDNA as described (28). Reactions containing 100 fmol of primed substrate and the common buffer in a final volume of 20 µl were preincubated at 37°C for 3 min. Synthesis was initiated upon addition of Pol κ or equivalent activity of Pol δ4. Aliquots were removed at 5, 15 and 30 min and quenched as described (28). Reaction products were separated on an 8% denaturing polyacrylamide gel and quantitated using a Molecular Dynamics Phosphoimager. The number of DNA molecules within three regions (5′ to the [GT]10 microsatellite, the [GT]10 microsatellite, and 3′ to the [GT]10 microsatellite) were determined by ImageQuant software and corrected for background and loading differences. [GT]10 termination probability is defined as the number of molecules within the [GT]10 microsatellite divided by the number of molecules within the [GT]10 microsatellite and 3′ to the [GT]10 microsatellite. Polymerase competition assays were performed similarly with 400 fmol of Pol δ4 ± 500 fmol PCNA, initiating synthesis in either moving (with 250 µM dNTPs) or stalled (-dATP) conditions. After 5 min, 40 fmol of Pol κ or an equivalent activity of Pol δ4 (200 fmol; thus, 600 fmol Pol δ4 final) was added (along with a final concentration of 250 µM dATP in the stalled reactions). Quantitation of reaction products was performed as above.
RESULTS
To measure the frequency and types of mutational events within repetitive DNA, we engineered the 5′-region of the HSV-tk gene to contain a variety of in-frame, short microsatellite (MS) sequences (Figure 1A). Gapped DNA heteroduplex molecules were constructed, and an in vitro DNA polymerase gap-filling assay was used to measure polymerase error rates within the MluI to StuI target (24). Forward mutations that inactivate the HSV-tk protein were scored after transfection of E. coli with the reaction products and selective plating (Figure 1B). An HSV-tk mutant phenotype can be generated either by mutations arising within the microsatellite motifs (that are not a multiple of three), or by mutations arising within the surrounding HSV-tk target coding sequence (Figure 1A). This experimental approach allows us to directly measure mutations in microsatellite alleles of varying sequence composition, and compare such events to mutations in the HSV-tk-coding sequence.
Replicative DNA polymerase fidelity within microsatellite alleles
We measured the fidelity of the recombinant 4-subunit holoenzyme form of human DNA polymerase δ (Pol δ4). Polymerase reactions were performed using gapped DNA substrates containing [GT]10, [TC]11 or [T]8 microsatellite reporter cassettes. The observed HSV-tk mutant frequencies for Pol δ4 were 8- to 44-fold higher than the ssDNA background frequency (Table 1). The observed HSV-tk mutant frequencies were adjusted for background and multiple mutational events after DNA sequence analyses to obtain the overall estimated polymerase error frequencies (overall Pol EFest; Table 1). This overall PolEFest is a result of polymerase errors within both the HSV-tk coding and microsatellite sequences of the target (Figure 1A). Polymerase error frequencies solely within the HSV-tk coding or the microsatellite sequences were calculated based on the proportion of mutational events within each of these regions. The majority (76–94%) of human Pol δ4 errors were produced within the microsatellite sequences for all three substrates (Table 1). The Pol EFest for Pol δ4 errors was high within the [GT]10 (2.3 × 10−3), [TC]11 (2.4 × 10−3), and [T]8 (3.1 × 10−3) alleles. We examined whether the PCNA/RFC complex could specifically increase Pol δ4 fidelity at microsatellite sequences. Pol δ4 fidelity within either the HSV-tk-coding sequences or the [GT]10 and [TC]11 microsatellites was not altered by the addition of PCNA and RFC (Table 1). An earlier study showed that these accessory proteins lower yeast Pol δ fidelity for base substitution errors (29).
Table 1.
Replicative DNA polymerase δ holoenzyme error rates within microsatellite and HSV-tk coding DNA sequences
| Polymerase | Observed HSV-tk frequencya ×10−4 (± SD) | Pol EFest × 10−4b (No. mutational events observed) |
||
|---|---|---|---|---|
| Overall | HSV-tk codingc | Microsatellitec | ||
| pSStu2 / [GT]10 template | ||||
| Noned | 4.1 | 0.26 (3) | <0.087 (0) | 0.26 (3) |
| Pol δ4 | 32 ± 5.2 | 27 (67) | 4.0 (10) | 23 (57) |
| Pol δ4 + PCNA/RFC | 41 | 31 (33) | 5.6 (6) | 25 (27) |
| pSStu4 / [TC]11 template | ||||
| None | 3.5 | 2.0 (26) | <0.078 (0) | 2.0 (26) |
| Pol δ4 | 43 ± 22 | 31 (62) | 7.5 (15) | 24 (47) |
| Pol δ4 + PCNA/RFC | 28 | 20 (24) | 6.7 (8) | 13 (16) |
| pSStu10/ [T]8 template | ||||
| None | 0.89 | ND | ND | ND |
| Pol δ4 | 39 ± 8.2 | 33 (32) | 2.1 (2) | 31 (30) |
aObserved mutant frequencies are mean of two, or mean ± SD of 3–6 independent reactions.
bPolymerase error frequency (Pol EF) was calculated using the equation: Pol EF = (Observed MF) – (ssDNA Background MF) – (Outside Target MF).
Pol EFs were adjusted for those mutants that had 2 or more mutational events in the target (see Methods). Mutants analyzed for DNA sequence changes were isolated from 2–3 independent polymerase reactions.
cCoding and microsatellite PolEFest were calculated by multiplying the proportion of mutational events at either site by the overall PolEFest.
dMutant frequency measured by electroporation of the ssDNA used to create the gapped DNA substrates. The corresponding overall Pol EF value is the frequency of mutations within the gapped target.
ND not determined.
Y-family DNA polymerase fidelity within microsatellites
We tested our hypothesis that microsatellite mutagenesis may vary with respect to DNA polymerase families, using human Y-family polymerases. Under our experimental buffer and template conditions, Pol κ and Pol η display similar or better DNA synthesis efficiencies to that of Pol δ4 (Supplementary Figure S3 and data not shown). The observed HSV-tk mutant frequencies for Pol κ and Pol η reactions were 3- to 16-fold higher than Pol δ4 for each template (Table 2). As a control, we compared the relative accuracy of the three polymerases during synthesis of the HSV-tk-coding sequences. The average coding Pol EFest was 4.5 × 10−4 for Pol δ4, 120 × 10−4 for Pol κ, and 380 × 10−4 for Pol η (Supplementary Table S1). Thus, Pol κ and Pol η are, on average, 27- and 84-fold less accurate than Pol δ4 at coding sequences, respectively. These data are in complete agreement with published results using the lacZ target and purified proteins from other sources (30–32).
Table 2.
Y Family DNA polymerase error rates within microsatellite and HSV-tk coding DNA sequences
| Polymerase | Observed HSV-tk frequencya × 10−4 (± SD) | Pol EFest × 10−4b (No. mutational events observed) |
||
|---|---|---|---|---|
| Overall | HSV-tk coding c | Microsatellite c | ||
| pSStu2 / [GT]10 template | ||||
| Pol κ | 140 ± 67 | 160 (160) | 140 (144) | 16 (16) |
| Pol η | 460 ± 52 | 410 (100) | 320 (79) | 86 (21) |
| pSStu4 / [TC]11 template | ||||
| Pol κ | 120 ± 39 | 100 (75) | 80 (60) | 20 (15) |
| Pol η | 680 ± 120 | 710 (130) | 430 (78) | 280 (52) |
| pSStu10 / [T]8 template | ||||
| Pol κ | 430 ± 85 | 460 (49) | 140 (15) | 320 (34) |
| Pol η | 430 ± 140 | 440 (35) | 210 (17) | 230 (18) |
aMutant frequencies are mean ± standard deviation of 3–6 independent reactions.
b,cSee legend to Table I for derivation of values.
Bold values indicate a low Pol κ error frequency for mutations arising within the [GT]10 and [TC]11 sequences.
When comparing polymerase fidelity at microsatellite sequences, we observed a low Pol κ error rate for mutations arising within the [GT]10 and [TC]11 dinucleotide sequences (Table 2). Remarkably, the Pol κ error frequency measured within dinucleotide alleles is 4- to 8-fold lower than the frequency of errors within the HSV-tk-coding region. Moreover, within these two microsatellite alleles, Pol κ does not display an error-prone phenotype, as the Pol EFest for Pol κ is not higher than that for Pol δ4 (Tables 1 and 2). This selective accuracy is specific for Pol κ and not a general feature of Y-family polymerases, since the Pol EFest for dinucleotide errors produced by Pol η is high for both microsatellite alleles (Table 2).
The creation of microsatellite interruptions by human DNA polymerases
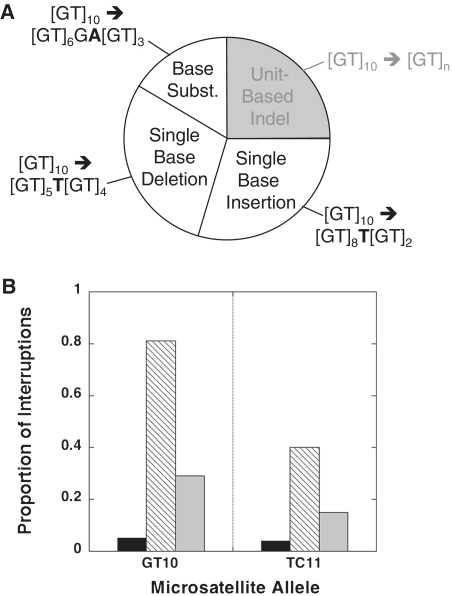
Various types of errors can be produced by polymerases within dinucleotide microsatellite alleles (33), and can be broadly classified into either unit-based insertion/deletion (indel) errors (where 2 nt is the unit size for dinucleotide repeats), or interruption errors (Figure 2A). Microsatellite unit-based indel errors characterize the mutational behavior of microsatellites observed in numerous model systems. If left uncorrected, such errors will result in allele length changes that can be a source of genomic instability (3). Interruption errors are any error that disrupts the tandem repeat sequence, converting the repetitive tract from one that is pure to one that is impure. Such errors in dinucleotide alleles can occur via multiple mechanisms, including single base indel or base substitution errors (Figure 2A). Although interruptions are polymerase errors, such errors are expected to be less detrimental genetically than unit-based indel errors. First, interruptions are known to stabilize expanded microsatellites (6). Second, microsatellite interruptions can divide the sequence into two smaller alleles with lengths below the threshold (34), thereby lowering mutability and initiating microsatellite degeneration.
Figure 2.
Specialized Y-family polymerases Pol κ and Pol η create more interruptions in dinucleotide microsatellites than does Pol δ4. (A) Types of mutations arising within dinucleotide microsatellites. Mutations within the dinucleotide microsatellite can occur via unit-based indel events (gray area) or by interruption events (white areas). Examples of interruption events by single base insertion, single base deletion, and base substitution mechanisms are shown for the [GT]10 allele. (B) Graphs depict the proportion of microsatellite interruptions among the total number of microsatellite mutational events at the [GT]10 and [TC]11 alleles for Pol δ4 (solid bars), Pol κ (hatched bars) and Pol η (gray bars).
We compared the propensity for the three polymerases to create interruption errors in vitro. The replicative Pol δ4 enzyme created very few interruptions within the alleles examined (Figure 2B). In stark contrast, Pol κ produced many interruption errors (up to 81%) within the GT10 and TC11 alleles, which is significantly different from Pol δ4 (P ≤ 0.0001 and P = 0.002, respectively, Fisher's exact test, Figure 2B). Thus, the Pol κ error frequency within the dinucleotide alleles is low (Table 2), and the few errors that are made by Pol κ are often interruptions, which may have beneficial consequences.
Distinct polymerase fidelity of unit-based indels within microsatellites
Both unit-based microsatellite allele length changes and traditional frameshift errors within coding sequences arise by a strand misalignment mechanism. Therefore, to directly compare the likelihood of the two types of polymerase misaligment errors, we calculated the Pol EFest for indel errors within the HSV-tk coding region versus unit-based indel errors within the microsatellite alleles.
The HSV-tk target sequence includes 23 ‘monitor’ sites of short tandem repeats (19 mononucleotide and four dinucleotide repeats of 2–3 units in length) that provide a good portrayal of polymerase indel errors occurring within coding sequences (34; underlined sequences in Supplementary Figure S2). As the HSV-tk coding region Pol EFest for each polymerase did not vary substantially between the different templates examined, a composite spectrum of coding region errors was created for each polymerase (Supplementary Figure S2). Strikingly, Pol κ produced primarily indel errors within monitor tandem repeats, as well as at non-repeated sequences. The Pol EFest for Pol κ or Pol η indel errors within the HSV-tk coding region is ∼100-fold higher than the Pol EFest for Pol δ4 (Figure 3A and Supplementary Table S1). These data establish that the hierarchy of polymerase accuracy for indel errors within short tandem repeats of coding sequences follows the order: Pol δ4 >> Pol κ or Pol η. A similar result was observed for the relative accuracy of Pols δ4, κ and η during synthesis of the [T]8 mononucleotide microsatellite allele. Specifically, Pols κ and η displayed a 10-fold higher Pol EFest for unit-based errors within the allele than did Pol δ4 (Figure 3B).
Figure 3.
Comparison of DNA polymerase unit-based indel accuracy. (A) HSV-tk-coding sequence monitors. For each polymerase, the coding region polymerase error frequencies (from Tables 1 and 2) were averaged among all templates, and the combined proportion of indel errors at the 23 monitor sites versus all detectable mutational events was calculated. This proportion was multiplied by the average coding PolEFest and the result graphed. One example of the type of mutations included in the analysis is depicted: the loss of one C residue within a short mononucleotide tandem repeat at HSV-tk positions 167–168. See Supplementary Figure S2 for complete representation of indel mutations at the 23 monitor sites. No mutational events were found at the coding region monitors among the mutants sequenced for the ssDNA background, and the error frequency is estimated to be <0.08 × 10−4. (B) [T]8 mononucleotide repeat. Indels at [T]8 are defined as any gain or loss of T units that change the length of the [T]8 tract to [T]n. The PolEFest for [T]8 indels was calculated by multiplying the proportion of MS unit indel errors by the MS PolEFest (Tables 1 and 2) for each polymerase. Background mutants from [T]8 ssDNA were not sequenced; therefore, the indel error frequency is ≤0.89 × 10−4. (C) [GT]10 microsatellite; and (D) [TC]11 microsatellite sequences. Indels at [GT]10 or [TC]11 are defined as any gain or loss of dinucleotide units that change the length of the tract. The PolEFest for [GT]10 and [TC]11 indels was calculated by multiplying the proportion of MS indel errors by the MS PolEFest (Tables 1 and 2) for each template and polymerase.
A very different hierarchy of polymerase fidelity was observed for unit-based indel errors within both dinucleotide alleles. Pol δ4 produced almost exclusively (89–95%) unit-based indel errors (and correspondingly few interruptions; Figure 2B), resulting in a high unit-based indel error frequency (Figure 3C and D). In contrast, few unit-based indel errors were observed in the Pol κ error spectra, because Pol κ creates many interruptions (Figure 2B). Thus, the resulting Pol EFest for unit-based indel errors produced by Pol κ was lower than that produced by Pol δ4 in both dinucleotide alleles (Figure 3C and D).
To compare polymerase indel error rates among targets of differing repeat lengths, we calculated the Pol EF/unit, where the unit size is one for mononucleotide and two for dinucleotide repeats (Table 3). For Pol δ4, the GT microsatellite indel error rate is 100-fold higher than HSV-tk-coding indel error rate, demonstrating the inherent inaccuracy of Pol δ4 at microsatellites (Table 3). Quite the opposite is true for Pol κ, as the ratio of the indel error rate within the GT microsatellite versus the HSV-tk-coding sequence is 0.17 (Table 3). Within the TC allele, the Pol δ4 indel error rate/unit is 22 × 10−5, and the Pol κ indel error rate/unit is 11 × 10−5. Thus, the ratio of polymerase error rates within the TC microsatellite/coding sequence is 130 for Pol δ4, but only 0.7 for Pol κ. The similarity in rank order of polymerase fidelity for GT and TC motifs suggests that Pol κ accuracy may be similar to or greater than that of replicative polymerases at other dinucleotide alleles. Our data demonstrate that the hierarchy of DNA polymerase fidelity is dependent upon DNA substrate sequence, and for dinucleotide microsatellites, follows the alternative order: Pol κ ≥ Pol δ4 > Pol η.
Table 3.
The hierarchy of DNA polymerase accuracy for indel errors is substrate-dependent
| Polymerase | Pol family | Indel polymerase error rate ×10−5 |
Ratio GT10 / HSV-tk | |
|---|---|---|---|---|
| HSV-tk codinga | [GT]10b | |||
| ypolεc | B | 0.033 | 9.0 | 270 |
| hpolδ | B | 0.17 | 22 | 130 |
| ypolδc | B | 0.23 | 25 | 110 |
| calf thymus polαd | B | 1.9 | 19 | 10 |
| rat polβe | X | 3.3 | 8.4 | 2.5 |
| hpolη | Y | 13 | 62 | 4.8 |
| hpolκ | Y | 18 | 3.0 | 0.17 |
aCoding indel monitor polymerase error rates are reported per unit and were determined by dividing the coding indel PolEFest by the total number of mono- and dinucleotide units included in all monitor sites (total from target sequence is 48).
b[GT]10 indel polymerase error rates are reported per unit and were determined by dividing the microsatellite indel PolEFest by the total number of GT units.
cData from (22).
dUnpublished data.
Cooperativity between Pol δ4 and Pol κ at dinucleotide microsatellites
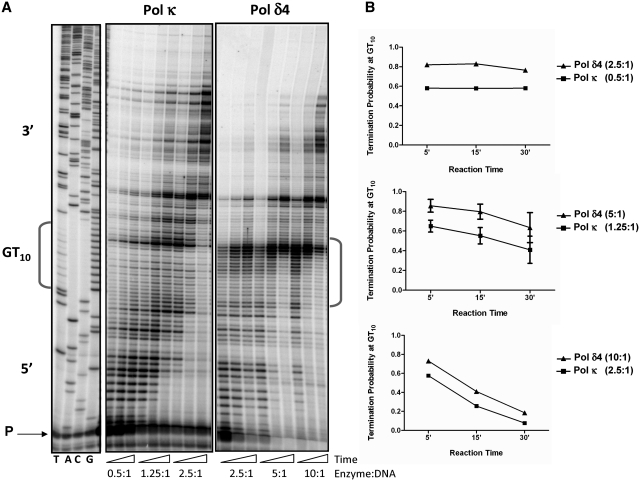
Using an E. coli model, Indiani et al. demonstrated that E. coli Pol IV can freely exchange with a moving replicative Pol III holoenzyme during DNA replication (35). In another model, an exchange between Saccharomyces cerevisiae Pol δ and Pol η was observed only upon stalling of the replicative polymerase (36). In both of these models, the polymerase exchange was monitored as a change in DNA synthesis rates and/or length of the DNA reaction products. Keeping these studies in mind, we ascertained whether Pol δ4 and Pol κ displayed polymerase-specific termination profiles within the GT10 microsatellite that could be used to monitor polymerase exchange. The termination probability of Pol δ4 within the GT allele was determined at three different polymerase:DNA template ratios (Figure 4). Under limiting enzyme conditions (2.5:1, Pol δ4:DNA ratio), we observed a high termination probability (0.76–0.84) within the [GT]10 sequence at all time points (Figure 4B, top panel). With higher amounts of enzyme, the termination probability decreased with increasing reaction time (Figure 4B, bottom panel), suggesting the product accumulation observed within the [GT]10 sequence is due to slowing of Pol δ4 synthesis rather than an absolute inhibition. To directly compare polymerase synthesis profiles, we determined the specific activities of our preparations of Pol δ4 and Pol κ using both primer extension (ssDNA template containing a [GT]10 sequence) and dTTP incorporation (poly dA/oligo dT primer-template) (Supplementary Figure S3). Under identical buffer conditions, Pol κ is 4- to 5-fold more active than Pol δ4. Therefore, we examined Pol κ termination probability within the [GT]10 sequence using 4–5-fold lower molar amounts of enzyme, relative to Pol δ4 (i.e. we compared equal activity). Under enzyme-limiting conditions (0.5:1, enzyme to DNA ratio), the Pol κ termination probability within the [GT]10 sequence was 0.56–0.60, lower than Pol δ4 at all time points (Figure 4B, top panel). With increasing Pol κ amounts, termination probability again decreased with reaction time. More importantly, however, the Pol κ termination probability was significantly lower than the Pol δ4 termination probability for every time point examined (P = 0.003, 5 and 15 min; P = 0.05, 30 min; unpaired student t-test) (Figure 4B, middle panel).
Figure 4.
Differential termination of Pol δ4 and Pol κ DNA synthesis within the [GT]10 microsatellite. (A) Representative phosphoimager scan of primer extension reaction products. The enzyme:DNA ratios for Pol κ (middle panel) and equivalent activity of Pol δ4 (right panel) are indicated. Triangles represent increasing reaction times (5, 15, 30 min). Reactions were run alongside a DNA sequencing ladder (left panel). (B) Quantitation of the reaction products. Termination probability was calculated by determining the number of molecules within the [GT]10 microsatellite divided by the number of molecules within and 3′ to the [GT]10 microsatellite. Graphs compare results from equal activities of Pol κ and Pol δ4. Data points are the mean of 2–6 independent reactions; error bars represent the standard deviation determined from 3 or more reactions.
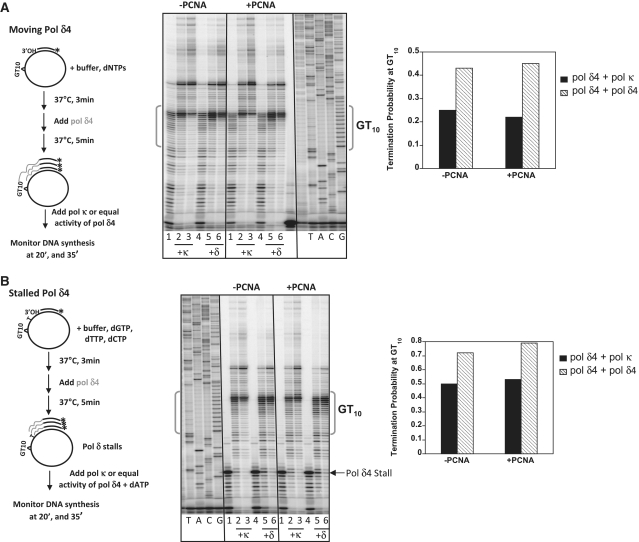
The differential termination observed above within the GT allele allowed us to test whether Pol κ could alleviate microsatellite-specific Pol δ4 termination. In preliminary experiments, we observed a significant decrease in GT10 termination when the synthesis reactions contained both Pol δ4 and Pol κ in similar molar amounts (0.5 pmol Pol δ4 and 0.25 pmol Pol κ), relative to reactions containing 0.75 pmol of Pol δ4 alone (Supplementary Figure S4). However, we also observed an appreciable increase in total primer-template extension in the dual polymerase reactions, relative to reactions with Pol δ4 alone. Because we also noted a difference in the specific activity of our two enzyme preparations (Supplementary Figure S3), we were concerned that using near equimolar polymerase amounts would give an unfair advantage to the more active polymerase preparation, in this case, Pol κ. Thus, we examined whether Pol κ could compete with Pol δ4 during microsatellite DNA synthesis using submolar amounts of Pol κ to control for specific activity differences. We examined two different experimental scenarios. In the first set of experiments, we allowed Pol δ4 to begin synthesis without any impediments, and then added either a 10-fold submolar amount of Pol κ, relative to Pol δ4, or an equal activity of additional Pol δ4 (as a control). After quantitating the resultant termination probability within the [GT]10 sequence, we observed that the amount of microsatellite-specific termination decreased when Pol κ was added to the reaction, relative to the Pol δ4 only reaction (Figure 5A). These results demonstrate that Pol κ can effectively compete with a moving Pol δ4 for primer-extension synthesis, even when Pol δ4 is present in a 10-fold molar excess over Pol κ. In the second set of experiments, we loaded Pol δ4 onto the DNA template in the presence of only three dNTP substrates in order to force Pol δ4 to stall prior to encountering the microsatellite sequence. Again, we added either Pol κ or an equal activity of additional Pol δ4, along with dATP, and analyzed DNA synthesis progression through the microsatellite. We observed a decrease of the microsatellite-specific Pol δ4 pause with addition of as little as 40 fmol Pol κ (Figure 5B). The presence of PCNA did not affect the results in either set of experiments, indicating that PCNA does not inhibit the competition between Pols δ4 and κ (Figure 5). These biochemical results suggest that Pols δ4 and κ can act cooperatively to reduce polymerase stalling within microsatellite sequences.
Figure 5.
Pol κ and Pol δ4 act cooperatively to alleviate pausing within the [GT]10 microsatellite. (A) Analyses performed under moving Pol δ4 conditions. Left panel: Schematic of the experimental design. Middle panel: Phosphoimager scan of reaction products obtained in either the absence (–PCNA) or presence (+PCNA) of 500 fmol PCNA. Lanes 1 and 4 of each reaction set show products after the 5 min incubation with 400 fmol Pol δ4 alone. Lanes 2 and 3 are products after addition of 40 fmol of Pol κ. The reactions were terminated after an additional 20 or 35 min. Lanes 5 and 6 are products after second addition of Pol δ4 (200 fmol for a total of 600 fmol) and termination after an additional 20 or 35 min. Polymerase conditions were chosen so that 100% of the available primer-template was extended by 20 min whether we added Pol κ or additional Pol δ4. Right panel: Termination probability within the [GT]10 sequence at 35 min under each polymerase condition. (B) Analyses performed under stalled Pol δ4 conditions. Pol δ4 stalling was created by excluding dATP during the first 5 min of the reaction. Reactions conditions are otherwise the same as in (A).
DISCUSSION
Intrinsic DNA features have been established as the primary determinants of microsatellite allele length variation (33). While the strand-slippage mechanism can readily account for the effects of microsatellite length (number of repeat units) on mutability, the mechanisms by which motif size and sequence composition affect microsatellite variation are less well established. In this study, we report that the human Y-family polymerase, Pol κ, displays a high accuracy during dinucleotide microsatellite DNA synthesis. Surprisingly, we observed that the relative DNA polymerase accuracy for unit-based indel errors during dinucleotide microsatellite DNA synthesis is: Pol κ ≥ Pol δ4 > Pol η (Figure 3C, D). This hierarchy is in distinct contrast to the one we observed within the HSV-tk-coding sequence control: Pol δ4 >> Pol κ or Pol η. Although Pol κ generated many strand-slippage errors within mononucleotide repeats, it is unusually accurate for unit-based indel events at the dinucleotide microsatellites (Figure 3). This behavior contrasts not only with human Pol δ4, but also with yeast Pol δ and Pol ε holoenzymes, and calf-thymus Polα-primase (Table 3). We measured a significantly higher termination probability for Pol δ4 than for Pol κ within the [GT]10 microsatellite (Figure 4), and used this differential termination to demonstrate that Pol κ can compete with a moving Pol δ4 enzyme during microsatellite DNA synthesis (Figure 5). These results demonstrate that Pol δ4 and Pol κ can cooperate enzymatically during dinucleotide microsatellite DNA synthesis. Our study conceptually extends the role of Pol κ beyond accurate translesion synthesis to include accurate synthesis of dinucleotide repetitive elements. As other studies (refs. 15–18) also have indicated a more widespread functional role of Y-family polymerases, we propose that the terminology ‘specialized polymerases’ to describe the Y-family polymerases may be more appropriate than the term TLS polymerases. Our data also demonstrate that the broad characterization of Y-family polymerases as error prone is not appropriate when considering the entire genome.
Pol κ could potentially promote microsatellite stability in two ways. First, this polymerase has an inherently high fidelity for unit-based indel errors within dinucleotide microsatellites (Figure 3C, D), and may act to limit microsatellite allele length variation. Second, when Pol κ does make errors within the microsatellites, it frequently creates interruption errors (Figure 2) within the long tandem repeat. We reported previously that a threshold length of 5 units exists for the mutability of GT/CA microsatellites in vitro and in human populations (34). Below the threshold length, the rate of mutation within the tandem repeat is not higher than the rate of indel mutations in coding sequences. Above the threshold, the mutability of the tandem repeat increases exponentially as the number of repeats increases. Therefore, the interruption errors made by Pol κ effectively break the long dinucleotide repeat arrays into two, shorter arrays that will mutate at a lower frequency. The in vitro results we report here are entirely consistent with the in vivo phenotype of Pol κ null mice. The spontaneous germline mutation rate within two expanded short tandem repeat (ESTR) alleles for Pol κ-deficient mice is significantly higher than within isogenic Pol κ wild-type mice (18).
All cells contain multiple DNA polymerases, and a widely held belief is that individual polymerases serve specialized functions that are necessary to maintain genome stability (19). The original paradigm for the functions of TLS polymerases posited that these enzymes promote the bypass of DNA lesions representing severe obstacles to replicative DNA polymerases, albeit at the cost of introducing mutations (37). This paradigm has shifted, as additional evidence has demonstrated that TLS polymerases can carry out error free bypass of specific DNA lesions in vivo, suggesting that this group of polymerases are structurally adapted for cognate lesions (38,39). Specifically, Pol κ has been implicated in efficient and error-free bypass in vivo of exocyclic N2-guanine lesions, such as benzo(a)pyrene diol epoxide formed by polycyclic aromatic hydrocarbons (PAH) (13,40).
The studies we report here extend this growing paradigm shift, with the novel finding that Pol κ is accurate when processing dinucleotide repetitive elements. The number of dinucleotide microsatellite alleles in the human reference genome (hg 18) is ∼227 000 for [GT/CA] alleles alone, and ∼500 000 for dinucleotide alleles of all sequence motifs (Supplementary Table S2). In comparison, the number of PAH adducts present in the genomes of mothers and newborns (41) or non-smokers (42) is 78–1080 adducts/cell, and rises to a high of 2040 adducts/cell in the lung epithelium of smokers (42). Clearly, the potential mutational load from polymerase errors during synthesis of microsatellites sequences in the human genome is much higher than that expected from PAH lesions. The exact role of Pol κ in genome replication will need to be investigated by additional in vivo studies.
The proposal that specialized polymerases, such as Pol κ, participate in chromosomal replication to maintain genomic integrity implies that genome-wide replication involves the cooperation of multiple DNA polymerases in addition to the classical replicative polymerases. In eukaryotes, a direct interaction has been shown between Rev1 and the Pol δ Pol32 subunit (43) as well as between Rev1 and Pol κ (44), consistent with models proposing that TLS polymerases may be an integral component of the replication fork. Indeed, human Pol κ can be found localized to PCNA-containing replication foci in undamaged cells (45). Current models of E. coli replication also propose the presence of multiple DNA polymerases at the replication fork. The active E. coli replisome has been shown by direct microscopy in living cells to contain three molecules of the replicative polymerase (46). In a biochemical study investigating the roles of Pol II and Pol IV outside of TLS, both polymerases were shown to have access to the DNA replication fork, and to freely exchange with Pol III in a moving replisome (35). The suggestion has been made that the third polymerase of the E. coli replisome may act as a reserve polymerase to assist during replication fork stalling, and that Pol II and Pol IV occupy the replisome to some extent during normal cellular growth (47). Thus, there exists the potential that specialized polymerases may encompass a role in replication at specific DNA structural elements. Our biochemical study (Figure 5) is similar to the previous analyses of replicative and Y-family DNA polymerase exchange (35,36,48), and extends this phenomenon to now include human Pol δ4 in addition to replicative polymerases of E. coli, T4 bacteriophage, and S. cerevisiae. Additionally, we are the first to provide evidence that Pol κ participates in an exchange reaction with Pol δ at an undamaged DNA sequence. We observed this exchange in the presence of a moving Pol δ4 enzyme, similar to what was observed for the E. coli Pol IV/Pol III system. Recently, Pol κ was shown to be present in a complex with Pol δ in human cells, and participate in DNA repair synthesis associated with nucleotide excision repair (17).
We presume that the differential accuracy of Pol κ and Pol δ4 within mononucleotide and dinucleotide repeats (Figure 3) reflects structural differences between the enzymes that affect utilization of bulged (misaligned) primer templates. Structural information is available for Pols κ and δ in complex with DNA and dNTP substrates (49,50), with the caveat that none of the structures to date correspond to the full length, wild-type (and for Pol δ, multisubunit) enzymes we have used in this study. The unique N-clasp interaction of Pol κ locks the polymerase around the DNA substrate, creating a restrictive active site (50). We found that Pol κ is accurate during synthesis of dinucleotide repeats, but inaccurate for mononucleotide repeats. Possibly, the intimate Pol κ interactions with the nascent basepair and unpaired nucleotides 5′ to the templating base may disfavor utilization of bulges greater than one nucleotide. Alternatively, the N-clasp may cause Pol κ to have a novel interaction with the templating strand, such that the enzyme does not sense the entirety of the long tandem dinucleotide repeat during DNA synthesis, but instead only reads the template one base at a time. This model would account for the unique pattern of interruption mutations (primarily, single base insertions or deletions) produced by Pol κ within the dinucleotide repeats. Pol δ, which primarily creates unit-based (two nucleotide) indels within the microsatellite, has a much larger protein footprint along the template strand through interactions of the N-terminal domain and the exonuclease domain (49). Such interactions of Pol δ may provide stabilizing contacts for looped out/bulged bases in the template strand, analogous to the stabilizing protein–extrahelical base interactions observed in the Dbh4 and Dpo4 polymerase-strand misalignment structures (51,52).
Microsatellite allele length variation, or microsatellite instability, is associated with 10–15% of colorectal, endometrial and gastric cancers, and has long been used as a diagnostic tool for Lynch Syndrome cancers. Alterations within microsatellites are generally accepted to be a consequence of strand slippage events during DNA replication that are left uncorrected by a defective mismatch repair system. Tumors of this type are characterized as MSI-High, with mononucleotide microsatellite markers proving to be more sensitive and predictive for Lynch Syndrome than dinucleotide markers (53). In addition, a group of sporadic colorectal carcinomas have been consistently identified, termed MSI-Low. The underlying cause of the MSI-Low phenotype is controversial, and it is unknown whether MSI-Low represents random mutations arising during clonal tumor cell evolution, or represents a mild mutator phenotype resulting from loss of a genome stability factor distinct from MMR (54–56). However, over- or under-expression of repair factors, such as alkyladenine DNA glycosylase (57) and methylguanine DNA methyltransferase (58), can contribute to an MSI-Low phenotype. Intriguingly, decreased Pol κ expression has been reported in colorectal tumors (59,60), relative to adjacent normal tissue. Results from our study and others may help to uncover unknown susceptibility factors in cancers exhibiting microsatellite instability.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
National Institutes of Health (grant numbers GM31973, ES014737 to M.Y.W.T.L., and CA100060, GM87472 to K.A.E.), and by generous contributions to the Gittlen Cancer Research Foundation of Penn State University. Funding for open access charge: National Institutes of Health (grant number GM87472 to K.A.E.)
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Mr. Guruprasad Ananda (Penn State University, Center for Comparative Genomics and Bioinformatics) for providing us with the human genome microsatellite enumeration data. We thank Drs. Joann Sweasy and Laura Carrel, and members of the Eckert laboratory for critical discussions and reading of the manuscript.
REFERENCES
- 1.Richard GF, Kerrest A, Dujon B. Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol. Mol. Biol. Rev. 2008;72:686–727. doi: 10.1128/MMBR.00011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Subramanian S, Mishra RK, Singh L. Genome-wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome biol. 2003;4:R13. doi: 10.1186/gb-2003-4-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gemayel R, Vinces MD, Legendre M, Verstrepen KJ. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu. Rev. Genet. 2010;44:445–477. doi: 10.1146/annurev-genet-072610-155046. [DOI] [PubMed] [Google Scholar]
- 4.Hefferon TW, Groman JD, Yurk CE, Cutting GR. A variable dinucleotide repeat in the CFTR gene contributes to phenotype diversity by forming RNA secondary structures that alter splicing. Proc. Natl Acad. Sci. USA. 2004;101:3504–3509. doi: 10.1073/pnas.0400182101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gangwal K, Sankar S, Hollenhorst PC, Kinsey M, Haroldsen SC, Shah AA, Boucher KM, Watkins WS, Jorde LB, Graves BJ, et al. Microsatellites as EWS/FLI response elements in Ewing's sarcoma. Proc. Natl Acad. Sci. USA. 2008;105:10149–10154. doi: 10.1073/pnas.0801073105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 7.Waters LS, Minesinger BK, Wiltrout ME, D'Souza S, Woodruff RV, Walker GC. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol. Mol. Biol. Rev. 2009;73:134–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sweasy JB, Lauper JM, Eckert KA. DNA polymerases and human diseases. Radiat. Res. 2006;166:693–714. doi: 10.1667/RR0706.1. [DOI] [PubMed] [Google Scholar]
- 9.Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, et al. The Y-family of DNA polymerases. Mol. Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 10.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 11.Johnson RE, Washington MT, Prakash S, Prakash L. Fidelity of human DNA polymerase eta. J. Biol. Chem. 2000;275:7447–7450. doi: 10.1074/jbc.275.11.7447. [DOI] [PubMed] [Google Scholar]
- 12.Masutani C, Kusumoto R, Iwai S, Hanaoka F. Mechanisms of accurate translesion synthesis by human DNA polymerase eta. Embo. J. 2000;19:3100–3109. doi: 10.1093/emboj/19.12.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Avkin S, Goldsmith M, Velasco-Miguel S, Geacintov N, Friedberg EC, Livneh Z. Quantitative analysis of translesion DNA synthesis across a benzo[a]pyrene-guanine adduct in mammalian cells: the role of DNA polymerase kappa. J. Biol. Chem. 2004;279:53298–53305. doi: 10.1074/jbc.M409155200. [DOI] [PubMed] [Google Scholar]
- 14.Jarosz DF, Godoy VG, Delaney JC, Essigmann JM, Walker GC. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature. 2006;439:225–228. doi: 10.1038/nature04318. [DOI] [PubMed] [Google Scholar]
- 15.Rey L, Sidorova JM, Puget N, Boudsocq F, Biard DS, Monnat RJ, Jr, Cazaux C, Hoffmann JS. Human DNA polymerase eta is required for common fragile site stability during unperturbed DNA replication. Mol. Cell. Biol. 2009;29:3344–3354. doi: 10.1128/MCB.00115-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Betous R, Rey L, Wang G, Pillaire MJ, Puget N, Selves J, Biard DS, Shin-ya K, Vasquez KM, Cazaux C, et al. Role of TLS DNA polymerases eta and kappa in processing naturally occurring structured DNA in human cells. Mol. Carcinog. 2009;48:369–378. doi: 10.1002/mc.20509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogi T, Limsirichaikul S, Overmeer RM, Volker M, Takenaka K, Cloney R, Nakazawa Y, Niimi A, Miki Y, Jaspers NG, et al. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol. Cell. 2010;37:714–727. doi: 10.1016/j.molcel.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 18.Burr KL, Velasco-Miguel S, Duvvuri VS, McDaniel LD, Friedberg EC, Dubrova YE. Elevated mutation rates in the germline of Polkappa mutant male mice. DNA Repair. 2006;5:860–862. doi: 10.1016/j.dnarep.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Bebenek K, Kunkel TA. Functions of DNA polymerases. Adv. Protein Chem. 2004;69:137–165. doi: 10.1016/S0065-3233(04)69005-X. [DOI] [PubMed] [Google Scholar]
- 20.Fortune JM, Pavlov YI, Welch CM, Johansson E, Burgers PM, Kunkel TA. Saccharomyces cerevisiae DNA polymerase delta: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J. Biol. Chem. 2005;280:29980–29987. doi: 10.1074/jbc.M505236200. [DOI] [PubMed] [Google Scholar]
- 21.Korona DA, Lecompte KG, Pursell ZF. The high fidelity and unique error signature of human DNA polymerase epsilon. Nucleic Acids Res. 2010;39:1763–1773. doi: 10.1093/nar/gkq1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abdulovic AL, Hile SE, Kunkel TA, Eckert KA. The in vitro fidelity of yeast DNA polymerase delta and polymerase varepsilon holoenzymes during dinucleotide microsatellite DNA synthesis. DNA Repair. 2011;10:497–505. doi: 10.1016/j.dnarep.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie B, Mazloum N, Liu L, Rahmeh A, Li H, Lee MY. Reconstitution and characterization of the human DNA polymerase delta four-subunit holoenzyme. Biochemistry. 2002;41:13133–13142. doi: 10.1021/bi0262707. [DOI] [PubMed] [Google Scholar]
- 24.Eckert KA, Mowery A, Hile SE. Misalignment-mediated DNA polymerase beta mutations: comparison of microsatellite and frame-shift error rates using a forward mutation assay. Biochemistry. 2002;41:10490–10498. doi: 10.1021/bi025918c. [DOI] [PubMed] [Google Scholar]
- 25.Hile SE, Eckert KA. DNA polymerase kappa produces interrupted mutations and displays polar pausing within mononucleotide microsatellite sequences. Nucleic Acids Res. 2008;36:688–696. doi: 10.1093/nar/gkm1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eckert KA, Hile SE, Vargo PL. Development and use of an in vitro HSV-tk forward mutation assay to study eukaryotic DNA polymerase processing of DNA alkyl lesions. Nucleic Acids Res. 1997;25:1450–1457. doi: 10.1093/nar/25.7.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Opresko PL, Sweasy JB, Eckert KA. The mutator form of polymerase beta with amino acid substitution at tyrosine 265 in the hinge region displays an increase in both base substitution and frame shift errors. Biochemistry. 1998;37:2111–2119. doi: 10.1021/bi9722711. [DOI] [PubMed] [Google Scholar]
- 28.Hile SE, Eckert KA. Positive correlation between DNA polymerase alpha-primase pausing and mutagenesis within polypyrimidine/polypurine microsatellite sequences. J. Mol. Biol. 2004;335:745–759. doi: 10.1016/j.jmb.2003.10.075. [DOI] [PubMed] [Google Scholar]
- 29.Hashimoto K, Shimizu K, Nakashima N, Sugino A. Fidelity of DNA polymerase delta holoenzyme from Saccharomyces cerevisiae: the sliding clamp proliferating cell nuclear antigen decreases its fidelity. Biochemistry. 2003;42:14207–14213. doi: 10.1021/bi0348359. [DOI] [PubMed] [Google Scholar]
- 30.Matsuda T, Bebenek K, Masutani C, Rogozin IB, Hanaoka F, Kunkel TA. Error rate and specificity of human and murine DNA polymerase eta. J. Mol. Biol. 2001;312:335–346. doi: 10.1006/jmbi.2001.4937. [DOI] [PubMed] [Google Scholar]
- 31.Ohashi E, Bebenek K, Matsuda T, Feaver WJ, Gerlach VL, Friedberg EC, Ohmori H, Kunkel TA. Fidelity and processivity of DNA synthesis by DNA polymerase kappa, the product of the human DINB1 gene. J. Biol. Chem. 2000;275:39678–39684. doi: 10.1074/jbc.M005309200. [DOI] [PubMed] [Google Scholar]
- 32.Schmitt MW, Matsumoto Y, Loeb LA. High fidelity and lesion bypass capability of human DNA polymerase delta. Biochimie. 2009;91:1163–1172. doi: 10.1016/j.biochi.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eckert KA, Hile SE. Every microsatellite is different: Intrinsic DNA features dictate mutagenesis of common microsatellites present in the human genome. Mol. Carcinog. 2009;48:379–388. doi: 10.1002/mc.20499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelkar YD, Strubczewski N, Hile SE, Chiaromonte F, Eckert KA, Makova KD. What is a microsatellite: a computational and experimental definition based upon repeat mutational behavior at A/T and GT/AC repeats. Genome Biol. Evol. 2010;2:620–635. doi: 10.1093/gbe/evq046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Indiani C, Langston LD, Yurieva O, Goodman MF, O'Donnell M. Translesion DNA polymerases remodel the replisome and alter the speed of the replicative helicase. Proc. Natl Acad. Sci. USA. 2009;106:6031–6038. doi: 10.1073/pnas.0901403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhuang Z, Johnson RE, Haracska L, Prakash L, Prakash S, Benkovic SJ. Regulation of polymerase exchange between Poleta and Poldelta by monoubiquitination of PCNA and the movement of DNA polymerase holoenzyme. Proc. Natl Acad. Sci. USA. 2008;105:5361–5366. doi: 10.1073/pnas.0801310105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo C, Kosarek-Stancel JN, Tang TS, Friedberg EC. Y-family DNA polymerases in mammalian cells. Cell. Mol. Life Sci. 2009;66:2363–2381. doi: 10.1007/s00018-009-0024-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Friedberg EC, Wagner R, Radman M. Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science. 2002;296:1627–1630. doi: 10.1126/science.1070236. [DOI] [PubMed] [Google Scholar]
- 39.Takata K, Wood RD. Bypass specialists operate together. EMBO J. 2009;28:313–314. doi: 10.1038/emboj.2008.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogi T, Shinkai Y, Tanaka K, Ohmori H. Pol kappa protects mammalian cells against the lethal and mutagenic effects of benzo[a]pyrene. Proc. Natl Acad. Sci. USA. 2002;99:15548–15553. doi: 10.1073/pnas.222377899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perera F, Hemminki K, Jedrychowski W, Whyatt R, Campbell U, Hsu Y, Santella R, Albertini R, O'Neill JP. In utero DNA damage from environmental pollution is associated with somatic gene mutation in newborns. Cancer Epidemiol. Biomarkers Prev. 2002;11:1134–1137. [PubMed] [Google Scholar]
- 42.Phillips DH, Hewer A, Martin CN, Garner RC, King MM. Correlation of DNA adduct levels in human lung with cigarette smoking. Nature. 1988;336:790–792. doi: 10.1038/336790a0. [DOI] [PubMed] [Google Scholar]
- 43.Acharya N, Johnson RE, Pages V, Prakash L, Prakash S. Yeast Rev1 protein promotes complex formation of DNA polymerase zeta with Pol32 subunit of DNA polymerase delta. Proc. Natl Acad. Sci. USA. 2009;106:9631–9636. doi: 10.1073/pnas.0902175106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohashi E, Hanafusa T, Kamei K, Song I, Tomida J, Hashimoto H, Vaziri C, Ohmori H. Identification of a novel REV1-interacting motif necessary for DNA polymerase kappa function. Genes Cells. 2009;14:101–111. doi: 10.1111/j.1365-2443.2008.01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bergoglio V, Bavoux C, Verbiest V, Hoffmann JS, Cazaux C. Localisation of human DNA polymerase kappa to replication foci. J. Cell Sci. 2002;115:4413–4418. doi: 10.1242/jcs.00162. [DOI] [PubMed] [Google Scholar]
- 46.Reyes-Lamothe R, Sherratt DJ, Leake MC. Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science. 2010;328:498–501. doi: 10.1126/science.1185757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langston LD, Indiani C, O'Donnell M. Whither the replisome: emerging perspectives on the dynamic nature of the DNA replication machinery. Cell Cycle. 2009;8:2686–2691. doi: 10.4161/cc.8.17.9390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang J, Zhuang Z, Roccasecca RM, Trakselis MA, Benkovic SJ. The dynamic processivity of the T4 DNA polymerase during replication. Proc. Natl Acad. Sci. USA. 2004;101:8289–8294. doi: 10.1073/pnas.0402625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase delta. Nat. Struct. Mol. Biol. 2009;16:979–986. doi: 10.1038/nsmb.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lone S, Townson SA, Uljon SN, Johnson RE, Brahma A, Nair DT, Prakash S, Prakash L, Aggarwal AK. Human DNA polymerase kappa encircles DNA: implications for mismatch extension and lesion bypass. Mol. Cell. 2007;25:601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 51.Wilson RC, Pata JD. Structural insights into the generation of single-base deletions by the Y family DNA polymerase dbh. Mol. Cell. 2008;29:767–779. doi: 10.1016/j.molcel.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 52.Wu Y, Wilson RC, Pata JD. The Y-family DNA polymerase Dpo4 uses a template slippage mechanism to create single-base deletions. J. Bacteriol. 2011;193:2630–2636. doi: 10.1128/JB.00012-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Imai K, Yamamoto H. Carcinogenesis and microsatellite instability: the interrelationship between genetics and epigenetics. Carcinogenesis. 2008;29:673–680. doi: 10.1093/carcin/bgm228. [DOI] [PubMed] [Google Scholar]
- 54.Halford S, Sasieni P, Rowan A, Wasan H, Bodmer W, Talbot I, Hawkins N, Ward R, Tomlinson I. Low-level microsatellite instability occurs in most colorectal cancers and is a nonrandomly distributed quantitative trait. Cancer Res. 2002;62:53–57. [PubMed] [Google Scholar]
- 55.Jass J, Whitehall V, Young J, Leggett B, Meltzer SJ, Matsubara N, Fishel R. Correspondance re: Laiho et al., Low level microsatellite instability in most colorectal carcinomas. Cancer Res. 2002;62:1166–1170. 5988–5990. [PubMed] [Google Scholar]
- 56.Laiho P, Launonen V, Lahermo P, Esteller M, Guo M, Herman JG, Mecklin JP, Jarvinen H, Sistonen P, Kim KM, et al. Low-level microsatellite instability in most colorectal carcinomas. Cancer Res. 2002;62:1166–1170. [PubMed] [Google Scholar]
- 57.Hofseth LJ, Khan MA, Ambrose M, Nikolayeva O, Xu-Welliver M, Kartalou M, Hussain SP, Roth RB, Zhou X, Mechanic LE, et al. The adaptive imbalance in base excision-repair enzymes generates microsatellite instability in chronic inflammation. J. Clin. Invest. 2003;112:1887–1894. doi: 10.1172/JCI19757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whitehall VL, Walsh MD, Young J, Leggett BA, Jass JR. Methylation of O-6-methylguanine DNA methyltransferase characterizes a subset of colorectal cancer with low-level DNA microsatellite instability. Cancer Res. 2001;61:827–830. [PubMed] [Google Scholar]
- 59.Lemee F, Bavoux C, Pillaire MJ, Bieth A, Machado CR, Pena SD, Guimbaud R, Selves J, Hoffmann JS, Cazaux C. Characterization of promoter regulatory elements involved in downexpression of the DNA polymerase kappa in colorectal cancer. Oncogene. 2007;26:3387–3394. doi: 10.1038/sj.onc.1210116. [DOI] [PubMed] [Google Scholar]
- 60.Pillaire MJ, Selves J, Gordien K, Gourraud PA, Gentil C, Danjoux M, Do C, Negre V, Bieth A, Guimbaud R, et al. A ‘DNA replication' signature of progression and negative outcome in colorectal cancer. Oncogene. 2010;29:876–887. doi: 10.1038/onc.2009.378. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.