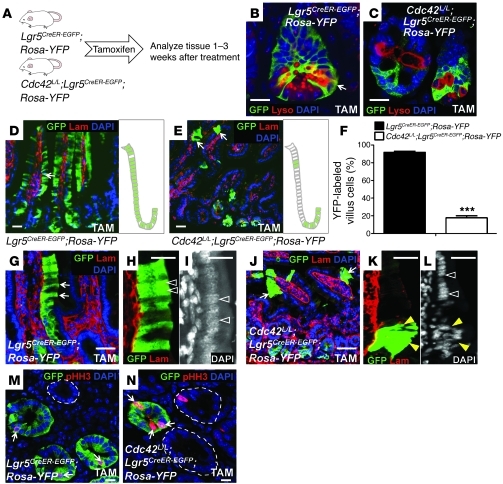
Figure 4. Cdc42 deficiency reduces the clonal expansion capacity of Lgr5 stem cells.
(A) Experimental scheme for the genetic tracing experiments. (B and C) GFP and lysozyme staining for control and Cdc42-deficient crypts 2 weeks after tamoxifen treatment. Arrow in B points to cells (in yellow) positive for both GFP and lysozyme. (D and E) GFP and basement membrane staining for control and Cdc42-deficient crypts 3 weeks after tamoxifen treatment. Arrows in E point to small clusters of cells derived from Cdc42-deficient stem cells. Schematic diagrams at the right of each panel summarize the results shown at left. (F) Quantification of RosaYFP-labeled villus epithelial cells derived from control and Cdc42-deficient Lgr5 stem cells 3 weeks after tamoxifen administration. ***P < 0.001. (G–L) GFP and basement membrane staining illustrate columnar shapes of labeled control intestinal epithelial cells (arrows in G) but abnormal morphology of labeled Cdc42-deficient cells (arrows in J). Arrowheads in H and K indicate positively labeled villus epithelial cells. White arrowheads in I and L indicate normal nuclear alignment. Yellow arrowheads in L indicate disrupted nuclear organization and cell polarity in villus cells derived from Cdc42-deficient stem cells. (M and N) Costaining of pHH3 and GFP. Dotted line in M indicates an unlabeled crypt. Dotted lines in N encircle the wild-type crypts that escaped Cre recombination. Arrows in M and N indicate GFP+/pHH3+ cells. Scale bars: 5 μm.