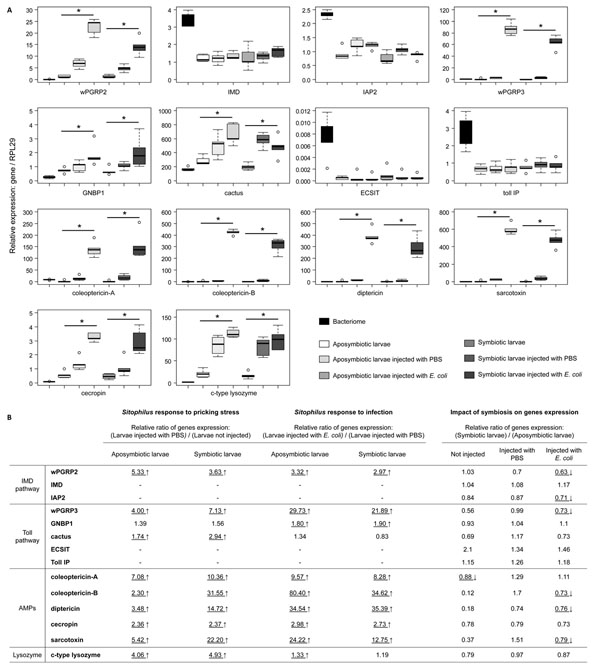
Figure 4.
Quantitative immune gene expression in symbiotic and aposymbiotic larvae of Sitophilus oryzae. (A) Transcript levels of immune genes quantified by qRT-PCR in whole aposymbiotic and symbiotic larvae. For both symbiotic and aposymbiotic larvae, non-injected larvae, larvae injected with PBS, and larvae injected with E. coli were analyzed. Results from gene expression in the bacteriome are reported here as an indicator. Represented expression of genes was normalized with the expression of the ribosomal protein L29. Each box represents the median (bolt line) and quartiles (25% / 75%) of five independent measurements. For each symbiotic and aposymbiotic status, the non-parametric Kruskal-Wallis test was applied in order to determine global difference between the three modalities tested (p-value < 0.05), represented by an asterisk. (B) Differential expression ratios obtained from q-RT-PCR experiments. For genes presenting significant differences in expression after the global test (see A), the pricking stress effect was tested by comparing larvae injected, or not, with PBS. The infection effect was tested by comparing larvae injected with PBS and larvae injected with E. coli. The REST pair-wise fixed reallocation randomization test was applied. For each modality tested (not injected, injected with PBS and injected with E. coli), a comparison between symbiotic larvae and aposymbiotic larvae was applied in order to evaluate the impact of symbiosis on the expression of immune genes. The REST pair-wise fixed reallocation randomization test was performed between the expression of genes from symbiotic and aposymbiotic larvae. Underlined scores indicate significant differences between the two modalities tested (p-value < 0.05). An up-arrow indicates upregulated genes whereas a down-arrow indicates downregulated genes.