Abstract
The immense biodiversity of the Atlas Mountains in North Africa might be the result of high rates of microallopatry caused by mountain barriers surpassing 4000 meters leading to patchy habitat distributions. We test the influence of geographic structures on the phylogenetic patterns among Buthus scorpions using mtDNA sequences. We sampled 91 individuals of the genus Buthus from 51 locations scattered around the Atlas Mountains (Antiatlas, High Atlas, Middle Atlas and Jebel Sahro). We sequenced 452 bp of the Cytochrome Oxidase I gene which proved to be highly variable within and among Buthus species. Our phylogenetic analysis yielded 12 distinct genetic groups one of which comprised three subgroups mostly in accordance with the orographic structure of the mountain systems. Main clades overlap with each other, while subclades are distributed parapatrically. Geographic structures likely acted as long-term barriers among populations causing restriction of gene flow and allowing for strong genetic differentiation. Thus, genetic structure and geographical distribution of genetic (sub)clusters follow the classical theory of allopatric differentiation where distinct groups evolve without range overlap until reproductive isolation and ecological differentiation has built up. Philopatry and low dispersal ability of Buthus scorpions are the likely causes for the observed strong genetic differentiation at this small geographic scale.
Introduction
The Mediterranean region comprises a highly diverse geographic area characterized by a variety of peninsulas and islands. This underlying geographic structure is reflected in biogeographical patterns of the region already described by de Lattin (1949) [1] who distinguished nine Mediterranean sub-centres. These early hypotheses have largely been confirmed by genetic analyses [2]. However, recent molecular data support even more fine-scaled patterns within these sub-centres; such patterns are especially known from the Iberian Peninsula e.g. [3], peninsular Italy e.g. [4] and the Balkans [5]. While the European part of the Mediterranean is biogeographically relatively well understood, the Maghreb still remains largely unstudied [6]. Therefore, the question emerges whether similar genetic substructures also exist in NW Africa.
De Lattin (1949) [1] suggested that the faunas of the Maghreb differ north and south of the High Atlas; this pattern is well reflected in the geographic distributions of many species e.g. [7]. On the contrary, other genetic studies revealed an east-west split along the Maghreb coast [8]. Furthermore, no genetic differentiation from Morocco to Tunisia was observed in the allozyme patterns of two butterfly species [9]. Nevertheless, all these genetic patterns are much simpler compared to findings in southern Europe. This is particularly astonishing considering the high orographic diversity of the Maghreb in general and the Atlas Mountains in particular. Yet, only few studies have addressed the phylo- or biogeographic patterns within the Atlas Mountains, although this area comprises an excellent region to study allopatric differentiation.
In order to test for the influence of orographic structures on differentiation and speciation processes, we selected the scorpion genus Buthus, which has undergone extensive speciation within NW Africa [10], as a model system. Buthus scorpions are widely distributed over large parts of Africa and show a circum-Mediterranean stronghold including the Mediterranean islands, with extraordinarily high diversity in North Africa [10]–[14]. Yet, molecular studies showed that diversity may still be underestimated and additional cryptic species may be abundant [14]. Most buthid scorpions are strictly territorial and philopatric and have low dispersal abilities. Consequently, individuals have limited home ranges which generally are restricted to small areas around their burrows [15]. Strong genetic differentiation has previously been observed for scorpion species and was probably triggered by barriers such as high elevations, steep valleys or sea barriers [16]–[19] in combination with low dispersal ability of the organisms [20]. In addition to being extremely limited, dispersal is also highly asymmetrical in these organisms, with male scorpions dispersing more widely. This apparently is a function of mate, prey and shelter search, where males exhibit a much more opportunistic locomotive behaviour, whereas females stay in their shelter and forage mainly close to the entrance of their burrows (“doorkeeping” strategy) [21], [22].
For this study we collected a total of 91 individuals belonging to 4 described Buthus species from 51 locations over major parts of the Antiatlas, High Atlas, Middle Atlas and Jebel Sahro during spring 2008 and 2009. Three specimens of the related genus Androctonus from two locations served as outgroup. We analysed a fragment of the mitochondrial Cytochrome Oxidase I (COI) gene, which has been shown to be highly informative at species level and consequently is one of the most commonly used markers for phylogenetic and phylogeographic studies. Due to its high potential for species identification it is often chosen as the barcoding gene in zoological studies. Here we use its properties to address the following questions:
Does genetic variation within Buthus exhibit geographic structuring?
Do genetic structures follow any geographical and/or evolutionary pattern?
Are genetic lineages in accordance with current taxonomy?
Results
We generated 94 sequences (including three outgroup specimens) with a total length of 452 base pairs. One sequence from Androctonus mauritanicus (JF820097) was obtained from genbank as an additional outgroup. Details about analyzed individuals, including genbank accession numbers, are given in Table 1. Out of the 452 sites analysed, 135 were polymorphic (29.9%) of which 127 were parsimony informative sites (4 sites had missing data). Within the ingroup we recovered 69 haplotype with a haplotypes diversity of Hd = 0.993 (SD = 0.003) and a nucleotide diversity of Pi = 0.0916 (SD = 0.0018). The average number of nucleotide differences was k = 41.138. Fst estimates among groups obtained from phylogenetic analyses were overall high and mostly significant (Table 2; non significant values are due to low sample sizes in some groups).
Table 1. Sampling localities of all studied Buthus populations.
| Group | Locality | Site | Coordinates(N; W) | Species | Date of sampling | Specimen ID/## | Genbank accession# |
| A | Tamensourt | 1 | 31.43; 8.01 | B. elmoutaouakili | 13-III-2009 | 9-80-5 | JN885948 |
| B | Chichoaua | 2 | 31.27; 8.47 | B. elmoutaouakili | 13-III-2009 | 9-79-1 | JN832012 |
| 9-79-2 | JN832013 | ||||||
| C | Argane | 3 | 30.55; 9.03 | B. elmoutaouakili | 12-III-2009 | 9-78-2 | JN832008 |
| 9-78-3 | JN832009 | ||||||
| D | Ameskrout | 4 | 30.37; 9.20 | B. elmoutaouakili | 12-III-2009 | 9-77-1 | JN832003 |
| 9-77-2 | JN832004 | ||||||
| Agadir | 5 | 30.28; 9.18 | B. elmoutaouakili | 12-III-2009 | 9-74-4 | JN831999 | |
| 9-74-6 | JN832000 | ||||||
| E | Tiznit | 6 | 29.18; 9.45 | B. elmoutaouakili | 24-II-2009 | 9-13-1 | JN831992 |
| 9-13-2 | JN831993 | ||||||
| Et Tnine | 7 | 29.42; 9.16 | B. elmoutaouakili | 11-V-2008 | 8-6-1 | JN831971 | |
| 8-6-2 | JN831972 | ||||||
| Tighermi | 8 | 29.32; 9.20 | B. elmoutaouakili | 24-II-2009 | 9-10-3 | JN831983 | |
| 9-10-5 | JN831984 | ||||||
| Onafka | 9 | 29.24; 9.15 | B. spec. | 24-II-2009 | 9-12-1 | JN831989 | |
| 9-12-2 | JN831990 | ||||||
| Izerbi | 10 | 29.20; 9.04 | B. spec. | 24-II-2009 | 9-11-1 | JN831986 | |
| 9-11-2 | JN831987 | ||||||
| F | Ait Saha | 11 | 30.06; 9.12 | B. elmoutaouakili | 23-II-2009 | 9-6-1 | JN831995 |
| 9-6-3 | JN831996 | ||||||
| Tiz-n-Test | 12 | 30.52; 8.23 | B. spec. | 12-V-2008 | 8-10-1 | JN831974 | |
| 8-10-2 | JN831975 | ||||||
| W Tassoumate | 13 | 30.30; 8.34 | B. elmoutaouakili | 11-III-2009 | 9-73-2 | JN832029 | |
| 9-73-3 | JN832030 | ||||||
| Ait Aissa | 14 | 30.18; 8.31 | Buthus spec. | 22-II-2009 | 9-3-1 | JN832016 | |
| 9-3-2 | JN832017 | ||||||
| Tassga | 15 | 30.10; 8.28 | Buthus spec. | 22-II-2009 | 9-5-1 | JN831978 | |
| 9-5-2 | JN831981 | ||||||
| S Tassoumate | 16 | 30.33; 8.15 | B. elmoutaouakili | 11-III-2009 | 9-72-1 | JN832026 | |
| 9-72-3 | JN832027 | ||||||
| Tassoumate | 17 | 30.35; 8.15 | B. elmoutaouakili | 11-III-2009 | 9-71-1 | JN832023 | |
| 9-71-2 | JN832024 | ||||||
| SW Taliouine | 18 | 30.22; 8.09 | B. elmoutaouakili | 11-III-2009 | 9-70-1 | JN831997 | |
| 9-70-3 | JN831998 | ||||||
| Igherm | 19 | 30.06; 8.70 | Buthus spec. | 22-II-2009 | 9-4-1 | JN832019 | |
| G | 75 km W Tazenakht | 20 | 30.30; 7.52 | B. malhommei | 10-III-2009 | 9-67-1 | JN885945 |
| 9-67-4 | JN885946 | ||||||
| 9-67-5 | JN885947 | ||||||
| W Tazenakht | 21 | 30.27; 7.39 | B. malhommei | 10-III-2009 | 9-66-2 | JN885943 | |
| 9-66-3 | JN885944 | ||||||
| Tazenakht | 22 | 30.28; 7.22 | B. malhommei | 10-III-2009 | 9-65-2 | JN885941 | |
| 9-65-5 | JN885942 | ||||||
| Tazenakht | 23 | 30.36; 7.16 | Buthus spec. | 28-II-2009 | 9-22-1 | JN885906 | |
| 9-22-2 | JN885907 | ||||||
| Agouine | 24 | 31.05; 7.17 | Buthus spec. | 3-III-2009 | 9-30-1 | JN885912 | |
| 9-30-2 | JN885913 | ||||||
| Iriri | 25 | 30.56; 7.13 | B. albengai | 28-II-2009 | 9-24-1 | JN885908 | |
| 9-24-2 | JN885909 | ||||||
| H | Aid Ben Haddou | 26 | 31.05; 7.08 | B. albengai | 1-III-2009 | 9-25-1 | JN885910 |
| 9-25-2 | JN885911 | ||||||
| Ouarzazate | 27 | 30.55; 6.51 | B. malhommei | 9-III-2009 | 9-62-2 | JN885939 | |
| 9-62-3 | JN885940 | ||||||
| I | Tata | 28 | 29.41; 8.09 | Buthus spec. | 25-II-2009 | 9-15-1 | JN885900 |
| 9-15-2 | JN885901 | ||||||
| Ouarzazate | 29 | 30.33; 7.09 | B. draa | 28-II-2009 | 9-21-1 | JN885904 | |
| 9-21-2 | JN885905 | ||||||
| Ouarzazate | 30 | 30.51; 6.51 | Buthus spec. | 4-III-2009 | 9-31-4 | JN885914 | |
| SE Ouarzazate | 31 | 30.48; 6.44 | Buthus spec. | 4-III-2009 | 9-32-4 | JN885915 | |
| Ait Sarin | 32 | 30.44; 6.38 | Buthus spec. | 4-III-2009 | 9-33-1 | JN885916 | |
| 9-33-2 | JN885917 | ||||||
| Rebat | 33 | 30.44; 6.26 | B. draa | 5-III-2009 | 9-34-3 | JN885918 | |
| 9-34-4 | JN885919 | ||||||
| Agdz | 34 | 30.41; 6.26 | B. draa | 5-III-2009 | 9-Agdz-1 | JN885956 | |
| 9-Agdz-2 | JN885957 | ||||||
| J | Fumzguid | 35 | 30.04; 6.52 | Buthus spec. | 27-II-2009 | 9-20-1 | JN885902 |
| 9-20-2 | JN885903 | ||||||
| K_a | Sidi-Flah | 36 | 31.00; 6.27 | B. malhommei | 9-III-2009 | 9-61-1 | JN885937 |
| 9-61-2 | JN885938 | ||||||
| Skoura | 37 | 31.01; 6.29 | B. malhommei | 9-III-2009 | 9-60-1 | JN885936 | |
| Skoura | 38 | 31.08; 6.21 | B. malhommei | 9-III-2009 | 9-59-5 | JN885935 | |
| M'Gouna | 39 | 31.14; 6.06 | B. malhommei | 9-III-2009 | 9-58-1 | JN885934 | |
| El-Kelaa | 40 | 31.17; 6.08 | B. malhommei | 8-III-2009 | 9-57-5 | JN885933 | |
| Boumalne de Dades | 41 | 31.22; 5.57 | B. malhommei | 8-III-2009 | 9-53-1 | JN885929 | |
| 9-53-2 | JN885930 | ||||||
| Juniter | 42 | 31.22; 5.47 | Buthus spec. | 7-III-2009 | 9-52-1 | JN885928 | |
| Emsoudar Aitsdrat | 43 | 31.27; 5.58 | B. malhommei | 8-III-2009 | 9-54-1 | JN885931 | |
| 9-54-3 | JN885932 | ||||||
| K_b | N'Kob | 44 | 30.51; 5.51 | B. draa | 4-III-2009 | 9-35-1 | JN885920 |
| Tazzarine | 45 | 30.50; 5.30 | B. draa | 5-III-2009 | 9-38-2 | JN885921 | |
| N Alnif | 46 | 31.10; 5.13 | B. draa | 5-III-2009 | 9-40-2 | JN885922 | |
| K_c | Tinerhir | 47 | 31.27; 5.36 | B. albengai | 7-III-2009 | 9-51-2 | JN885927 |
| Asselab | 48 | 31.32; 4.40 | B. draa | 6-III-2009 | 9-44-2 | JN885923 | |
| 9-44-3 | JN885924 | ||||||
| Ait El Farsi | 49 | 31.22; 5.17 | B. draa | 6-III-2009 | 9-47-1 | JN885925 | |
| 9-47-5 | JN885926 | ||||||
| Oulad Driss | 50 | 29.49; 5.38 | Buthus spec. | 16-III-2009 | 9-91-3 | JN885952 | |
| 9-91-6 | JN885953 | ||||||
| L | Draa Valley | 51 | 30.00; 5.32 | Buthus draa | 16-III-2009 | 9-A14-1 | JN885954 |
| 9-A14-4 | JN885955 | ||||||
| Out | Zagora | 52 | 33.85; 6.29 | Androctonus spec. | 17-III-2009 | 9-88-1 | JN885949 |
| Mesrate | 53 | 30.03; 5.36 | Androctonus spec. | 16-III-2009 | 9-89-1 | JN885950 | |
| 9-89-2 | JN885951 |
Given are the respective genetic groups, the locality names, a running number for each site (coinciding with figure 1), GPS coordinates of localities, morphological species assignment, date of captures and Genbank accession numbers.
Table 2. Fst values between populations estimated under the K2P substitution model; significance values (bold, α = 0.05) were assigned using 1000 permutations.
| A | B | C | D | E | F | G | H | I | J | K | L | |
| A | ||||||||||||
| B | 1.000 | |||||||||||
| C | 0.975 | 0.984 | ||||||||||
| D | 0.973 | 0.978 | 0.976 | |||||||||
| E | 0.881 | 0.903 | 0.901 | 0.884 | ||||||||
| F | 0.680 | 0.768 | 0.801 | 0.706 | 0.827 | |||||||
| G | 0.754 | 0.772 | 0.768 | 0.801 | 0.832 | 0.722 | ||||||
| H | 0.957 | 0.971 | 0.962 | 0.963 | 0.887 | 0.825 | 0.819 | |||||
| I | 0.813 | 0.842 | 0.832 | 0.821 | 0.750 | 0.789 | 0.801 | 0.841 | ||||
| J | 1.000 | 1.000 | 0.991 | 0.982 | 0.902 | 0.834 | 0.823 | 0.962 | 0.852 | |||
| K | 0.797 | 0.832 | 0.815 | 0.821 | 0.809 | 0.809 | 0.804 | 0.712 | 0.768 | 0.797 | ||
| L | 1.000 | 1.000 | 0.992 | 0.982 | 0.906 | 0.837 | 0.814 | 0.972 | 0.851 | 1.000 | 0.840 |
The second dataset included 84 additional sequences obtained from genbank (appendix SI). In total this more comprehensive dataset consisted of 179 sequences with 383 bp each. This included 148 variable sites (38.6%), 99 of which were parsimony informative. Many of these sites were uninformative due to a relatively large amount (60 sites) of missing data in genbank sequences.
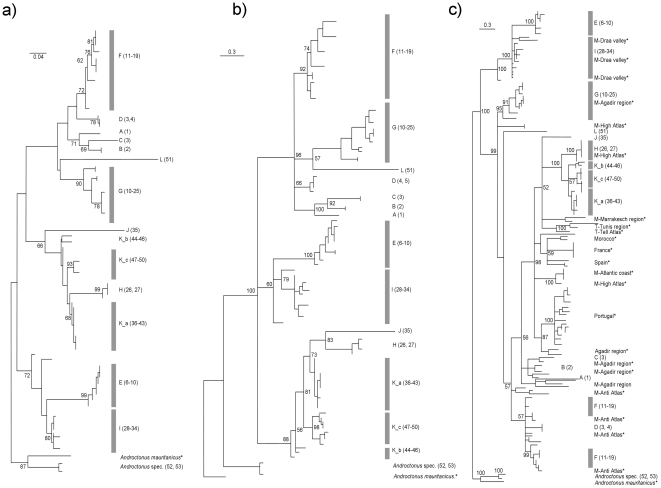
Reconstructed trees indicate the presence of 12 distinct Buthus lineages, which were recovered in all analyses (Fig. 1a–c). However, the relationships among groups differed depending on the tree reconstruction method. This is also reflected in the low statistical support for among group relationships. The support for most groups is high. The genetic patterns strongly coincide with the geographical distribution of the respective sampling locations. Lineages that occur sympatrically (K; most probably H/G/I, Figure 1) are genetically well separated in the trees, while genetically closer groups are all strictly allopatric or parapatric (e.g. I/E; H/K). The distributional patterns of most of the genetic clusters conform to the orographic pattern of the region (e.g. along valleys, southern or northern slopes of mountain systems).
Figure 1. Maximum Likelihood (ML) (a) and Bayesian Inference (BI) tree (b) rooted with Androctonus mauretanicus (N = 1) and Androctonus spec. (N = 3) as outgroups.
Numbers at nodes refer to bootstrap support for ML analyses and posterior probabilities for BI. Figure 1c shows a Bayesian Inference tree including all Buthus COI sequences available from genbank (sequences taken from genbank indicated by stars). For detailed information of sequences taken from genbank see appendix SI.
The comprehensive analyses of genbank data together with our own dataset generally yielded geographically meaningful clusters. The European samples represent two monophyletic groups, one in Portugal and one in France also including Spain. These clades likely correspond to the recently described species, Buthus montanus Lourenço & Vachon, 2004 and Buthus ibericus Lourenço & Vachon, 2004. However, the European Buthus are polyphyletic, yet restricted to one branch of the entire tree. They also make the North African Buthus paraphyletic (see Figure 1c).
The 12 lineages A through L do not match with current taxonomy (as reflected by morphology based identification keys): (i) B. elmoutaouakili Lourenço & Qui, 2006 is found in six lineages, B. malhommei Vachon, 1949 in four, B. albengai Lourenço, 2003 in two and B. draa Lourenço & Slimani, 2004 in four. None of the four species is monophyletic for our marker gene. (ii) Several lineages are composed of different species: four of the five lineages with ten or more individuals analysed contain two to three species; even lineage H (four individuals) includes two different morpho-types.
Discussion
The two tree hypotheses (ML and BI) constructed from our original dataset distinguish 12 genetic lineages. One of these clusters (lineage K) is further structured into three sub-clades. The genetic clusters exhibit a clear geographic pattern with all the lineages being confined to relatively restricted areas. None of the lineages is widely distributed. Most lineages are separated from each other by geographic barriers such as high mountain ranges or steep valleys. Only some basal lineages show range overlap (e.g. K) while all younger groups are strictly allopatric or parapatric (e.g. the three sub-groups of K). Four groups (A–D) are situated around the western and north-western parts of the High Atlas. One group is located in the westernmost Antiatlas (E), three others at the northern foothills of these mountains (F–H) and one at the southern foothills of the Antiatlas (J). The largest cluster (K) is distributed south of the High Atlas, east of the Draa valley, and splits further into three subclades, which form distinct strictly parapatric groups following the orographic structures of this region (e.g. along the Dades valley or restricted to the Jebel Sahro). Another cluster (I) extends from the Draa valley to the south western edge of the Antiatlas, probably including parts of the distribution ranges of clade G and H.
Barriers and retreats
The geographic distribution of the obtained genetic clusters strongly supports significant impact of orographic structures as dispersal barriers. The extant distribution of Buthus scorpions mostly does not exceed elevations higher than 2500 m asl [23], and individuals show low dispersal ability. Thus, high mountain ranges likely acted (and act) as strong barriers to gene flow resulting in genetically distinct clusters found within the Atlas Mountains. This is reflected in our data by fine structured genetic lineages of the sampled Buthus individuals. A similar pattern was observed for the same taxonomic group using various molecular markers such as allozymes, nuclear and mitochondrial DNA all of which showed significant genetic differentiation patterns even among local populations [24], [25]. Similar results were found for other organisms with low to moderate mobility [7]. These extremely sedentary species apparently diverged into several narrowly distributed genetic clusters, restricted to the slopes of mountains and river valleys (e.g. the Ourika and Draa). These lineages are found at different altitudes and might be adapted to various ecological niches. We assume that all actual distribution ranges of Buthus lineages also include their last centre of differentiation. Thus, our study area may comprise 12 allopatric refugia during the last ice age revealing a much more fine grained biogeographical sub-structure than previously observed in the Maghreb e.g. [7]–[9]. Consequently, the biogeographical structuring of Buthus in Morocco is much more complex than the structures observed in other Mediterranean sub-centres [26], most probably as a consequence of strong gene flow barriers represented by high mountain ranges and a more constant climatic situation in North Africa compared to the northern part of the Mediterranean. The combination of our data with previously analysed Buthus individuals (Figure 1c) underlines this superposition of the Maghreb region as a speciation and differentiation centre: The European Buthus are polyphyletic, but restricted to one branch of the entire tree and make the North African Buthus paraphyletic. This may be indicative of multiple colonisation events from North Africa to Europe.
The trigger of genetic differentiation
This distribution of genetic lineages supports an allopatric differentiation scenario as opposed to sympatric speciation processes [27]: The lineages originating from the oldest vicariance events are most strongly differentiated from each other. Perhaps this long period of time elapsed since the onset of differentiation allowed for the evolution of characteristics enabling sympatric occurrence of representatives of different genetic groups. Furthermore, the close geographic proximity among the well differentiated lineages G, H and I suggests some areas of range overlap among them. If reproductive barriers are in effect, species might even occur in sympatry without niche differentiation when resources are abundant. Thus, the time elapsed since the onset of the evolution of these lineages must have been sufficient that neither the inter-lineage competition for space nor the strong stenotopy of Buthus scorpions has been able to prevent the geographic intermixing of these lineages. However, differentiation among them still might not be sufficient for syntopic occurrences so that these groups exclude each other at each single locality. Such a scenario of competitive exclusion would reinforce a strictly geography-dependent distribution pattern of lineages.
Discordance between morphospecies and COI phylogeny
In contrast to the stringent geographic pattern, our findings are not congruent with the current taxonomic classification. The different species identified based on morphological characters often cluster within different genetic lineages which, in turn, often comprise more than one species. In addition, many specimens could not be identified due to incomplete or doubtful identification keys, and a lack of diagnostic traits. Hence, the current taxonomy of North African Buthus species is in need of revision. Additional diagnostic characters must be identified and new species keys be developed. However, to actually define the status of the detected genetic lineages, further morphological, behavioural and ecological studies are necessary. Besides, it is well known that gene trees and species trees may differ, e. g. due to shared ancestral polymorphisms, incomplete phylogenetic lineage sorting and introgression [28]. Evidence of recombination of mtDNA has been found in Buthus, which could also influence our results [29]. Taking our results as an exclusive basis for taxonomical conclusions would therefore doubtlessly be premature. In future studies, the addition of nuclear markers is indispensable to solve the question how prominent hybridization and introgression are among these Buthus lineages and species.
Materials and Methods
Sampling and identification
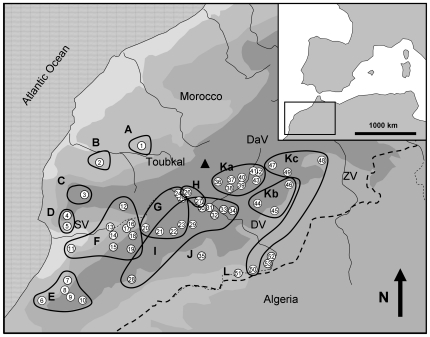
Most specimens were collected at daylight under stones. Overnight collections were facilitated by black light as all scorpions fluoresce in UV light due to a specific protein in their exoskeleton [30]. Collected specimens were stored in absolute ethanol until DNA extraction. All sampling sites are shown in Figure 2 and listed in Table 1 which also gives details on the sampling date and location. Species were identified with keys and additional species descriptions [10], [12], [31]–[33] based on typical morphological characters used for scorpion identification [cf. 32]–[33]. All species identifications were performed by the same author (IS).
Figure 2. Geographical distribution of all sampling sites and the 12 genetic groups and three subgroups around the Atlas Mountains in Morocco.
Numbers of sampling sites coincide with numbers given in Table 1. Letters of the groups correspond with Figure 1. The geographic coordinates of sampling sites are given in Table 1. Abbreviations: SV: Souss valley, DV: Draa valley, DaV: Dades valley, ZV: Ziz valley. Country border between Morocco and Algeria is indicated by a dotted line.
PCR and sequencing
For all individuals, DNA was isolated from leg, caudal segment or telson muscle tissue using the Qiagen DNeasy kit with standard protocols for tissue samples. A 452 bp fragment of the mitochondrial Cytochrome Oxidase I (COI) gene was amplified using standard PCR procedures with the following primers: forward 5′ GGT CAA CAA ATC ATA AAG ATA TTG G 3′, reverse 5′ TAA ACT TCA GGG TGA CCA AAA AAT CA 3′ [34]. PCRs were performed in 20 µl volumes: 10 µl Mastermix (Thermozyme), 0.2 µl of each Primer (1 µM), 4.6 µl PCR grade water and 5 µl DNA template. The cycle programme comprised an activation step at 94°C for 4 min, followed by 40 cycles of 30 sec denaturation at 94°C, 30 sec annealing at 45°C and 1 min elongation at 72°C. Cycling was terminated by a final extension step at 72°C for 10 min. Amplicons were subsequently purified and sequenced in both directions on an automated sequencer (3730xl DNA Analyzer; Applied Biosystems, Carlsbad, CA, USA) at the University of Kiel. All sequences are deposited at the NCBI genbank (appendix SI).
Data analysis
We included 91 sequences of Buthus, each 452 bp in length, in our analyses. Four sequences of the Buthid genus Androctonus served as outgroup (one sequence from A. mauretanicus obtained from genbank: accession JF820097 and three sequences from Androctonus spec. (09-88-1; 09-89-1; 09-89-2)). Sequences were inspected and aligned using Geneious 5.0.3 [35].
General alignment statistics (diversity indices, etc.) were calculated using DNAsp v. 5.10 [36]. Estimates of population divergence were calculated as pairwise Fst under a Kimura2P substitution model in Arlequin v.3.5.1.2. [37]. Populations were defined a posteriori according to our phylogenetic results. Significance of Fst values was tested using 1000 permutations and a significance level of 0.05.
The best fitting model of sequence evolution for phylogenetic tree reconstruction was chosen using MrModeltest 2.3 [38] in PAUP 4.0 b10 [39] and was determined to be the GTR+I+G. Maximum Likelihood (ML) trees were constructed using MEGA version 5.01 [40]; 1000 bootstrap replicates were performed to obtain a statistical measure for branch support (Figure 1a). Additionally, we used the Bayesian algorithm implemented in MrBayes 3.1.2 [41] to obtain a second phylogenetic inference and evaluate the stability of our tree reconstruction (Figure 1b). Using the obtained substitution model we ran the Markov chain Monte Carlo algorithm with four chains and two independent runs for 20 million generations. Trees were sampled every 2,000 generations; the first 10% of generated trees were discarded as burn-in as recommended by Tracer v1.5 [42]. The remaining trees were used to generate a consensus tree.
In a second analysis, we included 84 sequences from Buthus scorpions obtained from genbank (see appendix SI) resulting in a total of 175 ingroup sequences. However, due to only partial overlap the alignment was reduced to 383 bp in length. Bayesian analyses of this dataset were performed as described above. While these additional analyses provide further insights into geographic structuring of genetic divergence, they do not help in evaluating species identity and monophyly (Figure 1c). When these sequences were submitted to Genbank, the vast majority of Moroccan Buthus species known today had not yet been described and most Moroccan Buthus were characterized as subspecies of B. occitanus (B. o. occitanus – Europe, B. o. mardochei – Morocco, B. o. tunetanus – Tunisia) [31].
Supporting Information
DNA sequences obtained from NCBI genbank.
(DOC)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Funding was performed by the “Fonds National de la Recherche Luxembourg” and the “German Academic Exchange Service”; furthermore, the Natural History Museum Luxembourg supported field work and laboratory analyses. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.de Lattin G. Beiträge zur Zoogeographie des Mittelmeergebietes. Verhandlungen der Deutschen Zoologischen Gesellschaft Kiel. 1949;1948:143–151. [Google Scholar]
- 2.Hewitt GM. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- 3.Gómez A, Lunt DH. Weiss S, Ferrand N, editors. Refugia within refugia: patterns of phylogeographic concordance in the Iberian Peninsula, Phylogeography of southern European refugia. 2007. pp. 155–188. Springer, Heidelberg.
- 4.Joger U, Fritz U, Guicking D, Tripepi S, Pennisi MG, et al. Phylogeography of western Palaearctic reptiles - Spatial and temporal speciation patterns. Zoologischer Anzeiger. 2007;246:293–313. [Google Scholar]
- 5.Schmitt T, Habel JC, Zimmermann M, Müller P. Genetic differentiation of the Marbled White butterfly Melanargia galathea accounts for glacial distribution patterns and postglacial range expansion in south-eastern Europe. Mol Ecol. 2006;15:1889–1901. doi: 10.1111/j.1365-294X.2006.02900.x. [DOI] [PubMed] [Google Scholar]
- 6.Hewitt GM. Genetic consequences of climatic oscillation in the Quaternary. Phil Trans Roy Soc Lond. 2004;359:183–195. doi: 10.1098/rstb.2003.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fritz U, Barata M, Busack SD, Fritzsch G, Astilho R. Impact of mountain chains, sea straits and peripheral populations on genetic and taxonomic structure of a freshwater turtle, Mauremys leprosa (Reptilia, Testudines, Geoemydidae). Zool Sci. 2006;35:97–108. [Google Scholar]
- 8.Cosson JF, Hutterer R, Libois R, Sarà M, Taberlet P, et al. Phylogeographical footprints of the Strait of Gibraltar and quaternary climatic fluctuations in the western Mediterranean: a case study with the greater white-toothed shrew Crocidura russula (Mammalia: Soricidae). Mol Ecol. 2005;14:1151–1162. doi: 10.1111/j.1365-294X.2005.02476.x. [DOI] [PubMed] [Google Scholar]
- 9.Habel JC, Rödder D, Scalercio S, Meyer M, Schmitt T. Strong genetic cohesiveness between Italy and the Maghreb in four butterfly species. Biol J Linn Soc. 2010;99:818–830. [Google Scholar]
- 10.Lourenço W. Additions to the scorpion fauna (Arachnida) of North Africa with special reference to the genus Buthus Leach, 1815. Revue Suisse de Zoologie. 2003;110:875–912. [Google Scholar]
- 11.Fet V, Sissom WD, Lowe G, Braunwalder ME. Catalog of the Scorpions of the World (1758–1998). 2000. The New York Entomological Society, New York.
- 12.Stockmann R, Ythier E. Scorpions of the World. 2010. 568 N.A.P. Editions. France.
- 13.Lourenco WR. Considerations sur les modeles de distribution et differentiation du genre Buthus Leach, 1815, avec la description d'une nouvelle espece des montagnes du Tassili des Ajjer, Algerie (Scorpiones, Buthidae). Biogeographica. 2002;78:109–127. [Google Scholar]
- 14.Gantenbein B, Keightley PD. Rates of molecular evolution in nuclear genes of east Mediterranean scorpions. Evolution. 2004;58:2486–2497. [PubMed] [Google Scholar]
- 15.Polis GA, Sissom WD. Life history, In: Polis GA, editor. The biology of scorpions. Stanford, CA: Stanford University Press; 1990. pp. 161–22. [Google Scholar]
- 16.Froufe E, Sousa P, Alves PC, Harris DJ. Genetic diversity within Scorpio maurus (Scorpiones: Scorpionidae) from Morocco: Preliminary evidence based on CO1 mitochondrial DNA sequences. Biologia. 2008;63:1157–1160. [Google Scholar]
- 17.Gantenbein B, Largiadièr CR. The phylogeographic importance of the Strait of Gibraltar as a gene flow barrier in terrestrial arthropods: a case study with the scorpion Buthus occitanus as model organism. Mol Phyl Evol. 2003;28:119–130. doi: 10.1016/s1055-7903(03)00031-9. [DOI] [PubMed] [Google Scholar]
- 18.Sousa P, Froufe E, Alves PC, Harris DJ. Genetic diversity within scorpions of the genus Buthus from the Iberian Peninsula: mitochondrial DNA sequence data indicate additional distinct cryptic lineages. J Arachnol. 2010;38:206–211. [Google Scholar]
- 19.Sousa P, Froufe E, Harris DJ, Alves PC, van der Meijden A. Genetic diversity of Maghrebian Hottentotta (Scorpiones: Buthidae) scorpions based on CO1: new insights on the genus phylogeny and distribution. African Invertebrates. 2011;52:135–143. [Google Scholar]
- 20.Polis GA, McReynolds CN, Glenn Ford R. Home range geometry of the desert scorpion Paruroctonus mesaensis. Oecologia. 1985;67:273–277. doi: 10.1007/BF00384298. [DOI] [PubMed] [Google Scholar]
- 21.Kaltsas D, Mylonas M. Locomotory activity and orientation of Mesobuthus gibbosus (Scorpiones: Buthidae) in central Aegean Archipelago. J Nat Hist. 2010;44:1445–1459. [Google Scholar]
- 22.Kaltsas D, Stathi I, Mylonas M. The foraging activity of Mesobuthus gibbosus (Scorpiones: Buthidae) in central and south Aegean archipelago. J Nat Hist. 2008;42:513–527. [Google Scholar]
- 23.Brownell PH, Polis GA. Brownell PH, Polis GA, editors. Scorpion biology and research, 2001. 448 Oxford University Press. Oxford.
- 24.Yamashita T, Polis GA. A test of the central-marginal model using sand scorpion populations (Paruroctonus mesaensis). J Arachnol. 1995;23:60–64. [Google Scholar]
- 25.Gantenbein B. The genetic population structure of Buthus occitanus (Scorpiones: Buthidae) across the Strait of Gibraltar: calibrating a molecular clock using nuclear allozyme variation. Biol J Linn Soc. 2004;81:519–534. [Google Scholar]
- 26.Schmitt Molecular biogeography of Europe: Pleistocene cycles and postglacial trends. Front Zool. 2007;4:11. doi: 10.1186/1742-9994-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barraclough TG, Vogler AP. Detecting the Geographical Pattern of Speciation from Species-Level Phylogenies. Am Nat. 2000;155:419–434. doi: 10.1086/303332. [DOI] [PubMed] [Google Scholar]
- 28.Pollard DA, Iyer VN, Moses AM, Eisen MB. Widespread Discordance of Gene Trees with Species Tree in Drosophila: Evidence for Incomplete Lineage Sorting. PLoS Genetics. 2006;2:10. doi: 10.1371/journal.pgen.0020173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gantenbein B, Fet V, Gantenbein-Ritter IA, Balloux F. Evidence for recombination in scorpion mitochondrial DNA (Scorpiones: Buthidae). Proc Roy Soc Biol Sci. 2005;272:697–704. doi: 10.1098/rspb.2004.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anglade F, Ricordel I, Goyffon M. Données spectroscopiques sur la fluorescence de la cuticule de scorpion. Bull Soc Europ Arachnol. 1990;1:5–9. [Google Scholar]
- 31.Vachon M. Genre Buthus. Etudes sur les Scorpions. 1952. pp. 241–316. Institute Pasteur D' Algerie. Alger.
- 32.Vachon M. Remarques sur l'utilisation en systématique des soies sensorielles (trichobotries) chez les scorpions du genre Euscorpius Thorel (Chactidae). Bulletin du Musée National d'Histoire Naturelles (Paris) Série . 1962;2: 34:345–354. [Google Scholar]
- 33.Vachon M. Remarques sur l'utilisation en systématique des soies sensorielles (trichobotries) chez les scorpions du genre Euscorpius Thorel (Chactidae). Bulletin du Musée National d'Histoire Naturelles (Paris) 1962;34:345–354. [Google Scholar]
- 34.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotech. 1994;3:294–299. [PubMed] [Google Scholar]
- 35.Drummond AJ, Kearse M, Heled J, Moir R, Thierer T, et al. Geneious v5.0.3. Geneious website. 2006. Available: www.geneious.com. Accessed 2011 Feb 31.
- 36.Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 37.Excoffier L, Lischer HEL. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Res. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 38.Nylander JAA. MrModeltest 2.3. MrModeltest website. 2004. Available: www.abc.se/nylander/. Accessed 2011 Feb 31.
- 39.Swofford DL. 1991. PAUP: Phylogenetic Analysis Using Parsimony, Version 3.1 Computer program distributed by the Illinois Natural History Survey, Champaign, Illinois.
- 40.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huelsenbeck JP, Ronquist R. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 42.Rambaut A, Drummond AJ. Tracer Release 1.4. Tracer Release website. 2007. Available: www.beast.bioed.acuk/Tracer. Accessed 2011 Feb 31.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DNA sequences obtained from NCBI genbank.
(DOC)