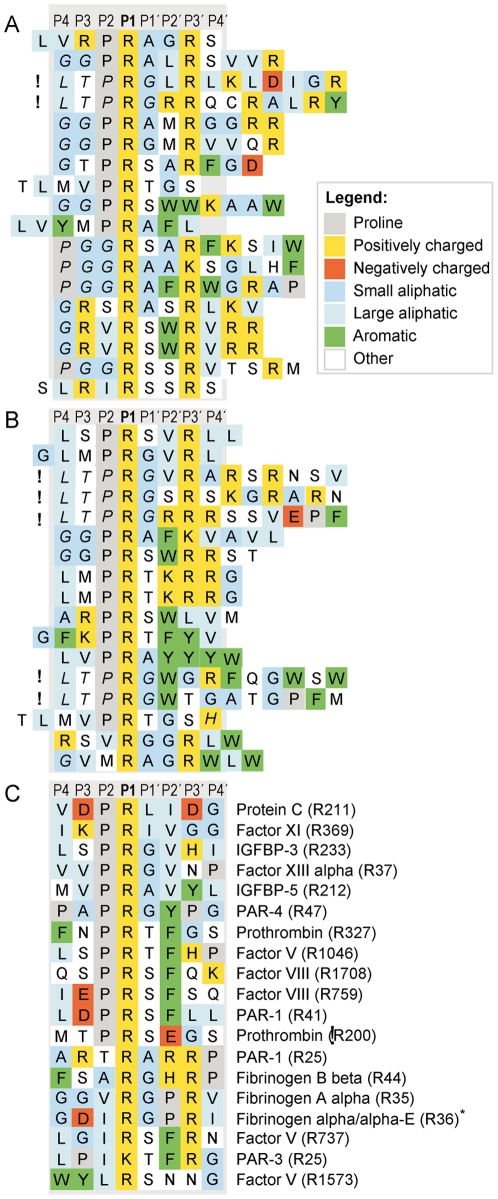
Figure 1. Alignment of sequences obtained after five selection rounds with 1 U of thrombin or 0.2 U of thrombin, compared to natural substrates.
Panel A shows the result with 1 U of thrombin, panel B the result with 0.2 U of thrombin and panel C a panel of natural substrates. The P1 residue in natural substrates (after which cleavage occurs) is denoted in parentheses. Substrate sequences refer to Homo sapiens where not indicated otherwise.! marks phage sequences that have LTPRG instead of LTPGG in the N-terminal flank. Residues from the non-randomized phage region are in italics. *, from Rattus norvegicus; IGFBP, insulin-like growth factor-binding protein; PAR, protease-activated receptor. The cleavage site of thrombin in the natural substrates listed in panel C is numbered from the N terminal of the pre-pro protein, from the first methionine. This list of natural substrates is a selection of a few of the most well known substrates of this enzyme. However, the list of potential in vivo substrates is much longer and includes many other proteins such as protein S, TAFI, antithrombin, heparin cofactor II and nexin I.