Abstract
Development of specific immunotherapy for colorectal cancer (CRC) will require identification of antigens selectively or exclusively expressed on CRC cells and strategies to induce and enhance immune responses against these antigenic targets. Cancer-testis (C-T) antigens are proving to be excellent targets for immunotherapy of solid tumors such as melanoma, but their clinical utility for treatment of CRC has to date been limited by their infrequent expression in CRC cells. Here we report that the hypomethylating agent 5-aza-2′-deoxycytidine (DAC) induces expression of NY-ESO-1 and other C-T genes in CRC cells both in vitro and in vivo in a dose-dependent manner but has negligible effects on the expression of C-T genes in normal non-transformed cells such as fibroblasts. The induction by DAC of NY-ESO-1 expression in CRC cells persists over 100 days after DAC exposure and is associated with increased levels of NY-ESO-1 protein. CRC cells exposed to DAC at concentrations that can be readily achieved in vivo are rendered susceptible to MHC-restricted recognition by CD8+ NY-ESO-1-specific T cells. We also demonstrate that retroviral transduction of polyclonal peripheral blood T cells from a metastatic CRC patient with the T cell receptor (TCR) α and β chain genes encoding an HLA-A2-restricted, NY-ESO-1157-165-specific TCR can be used to generate both CD8+ and CD4+ NY-ESO-1157-165-specific T cells that selectively recognize DAC-treated CRC but not non-transformed cells. Collectively, these results suggest that the combination of epigenetic modulation and adoptive transfer of genetically engineered T lymphocytes may enable specific immunotherapy of CRC.
Keywords: cancer-testis antigens, colorectal cancer, NY-ESO-1, cytolytic T lymphocyte, real-time cell electrical sensing assay
INTRODUCTION
Colorectal cancer (CRC) is one of the most common malignancies and the third leading cause of cancer-related death worldwide.1 Despite modest recent advances in systemic therapies, the majority of patients with advanced or metastatic CRC ultimately die from their disease. There is evidence that the population of T lymphocytes, and in particular the CD8+ cytotoxic T lymphocytes (CTLs), that infiltrates CRC tumors influences the natural history of the disease,2-9 suggesting the existence of an antigen-specific tumor-reactive immune response. Identification of suitable target antigens that are commonly expressed on CRC cells could enable the development of antigen-specific T cell therapy for CRC.
Adoptive T cell therapy has shown promise for the treatment of selected solid tumors such as metastatic melanoma.10-15 However, the development of T cell therapy for CRC has been slowed, in part, by a paucity of suitable target antigens. The cancer-testis (C-T) antigens comprise a growing class of more than one hundred immunogenic proteins and protein families that are expressed in germ cells, trophoblastic tissues, and a wide range of cancers, but not in normal somatic tissues.16 Given their restricted expression and immunogenicity, they are attractive targets for immunotherapy. However, expression of C-T antigens is infrequent in most CRC tumors.17, 18 Silencing of C-T genes in some cancer cells is commonly mediated by methylation of CpG islands near the promoter of C-T genes. The DNA methylation inhibitor 5-aza-2′-deoxycytidine (DAC) induces C-T gene expression in a variety of cancer types, but this effect has not been extensively studied in CRC to date.
The prototypic C-T antigen NY-ESO-1 has emerged as one of the most attractive target antigens for immunotherapy because it frequently induces CD4+ and CD8+ T cell responses as well as antibody responses in cancer patients whose tumors express it, and it is expressed in a significant proportion of cancer cells of diverse histology.19 Expression of NY-ESO-1 can also be selectively induced by DAC in transformed, but not non-transformed, cells that do not natively express it. DAC-induced expression of NY-ESO-1 in a diverse group of cancers with varying histologies has been correlated with recognition by NY-ESO-1-specific CTL clones.20-25 Previous studies have reported induction by DAC of NY-ESO-1 expression in a few CRC cell lines.21, 25 but the effect of DAC exposure on recognition of CRC cells by C-T antigen-specific CTL has not been extensively explored. We therefore investigated whether DAC could induce expression of NY-ESO-1 and other C-T genes in CRC cell lines and also sensitize them to NY-ESO-1-specific immunotherapy.
A minority of CRC patients show evidence of spontaneous immunity to NY-ESO-1,17 and it is anticipated that autologous NY-ESO-1-specific T cells would not be available for all CRC patients who might otherwise be eligible for NY-ESO-1-specific T cell therapy. Autologous NY-ESO-1-specific T cells suitable for use in adoptive therapy have been generated through transfer of the T cell receptor (TCR) α and β chain genes from a high affinity NY-ESO-1-specific CTL clone (termed 1G4) into polyclonal peripheral blood mononuclear cells (PBMC).15, 24, 26-28 These gene-modified T cells expressing transgenic 1G4 TCR exert potent in vitro effector function against a variety of cancer cell lines including those in which NY-ESO-1 expression has been induced by DAC.24 In a recent clinical trial in which adoptive transfer of genetically modified T cells expressing a high affinity mutant of the 1G4 TCR into melanoma and synovial sarcoma patients whose tumors expressed NY-ESO-1, objective tumor regression was observed in several patients in the absence of significant toxicity.15 We therefore investigated whether this promising method of generating NY-ESO-1-specific T cells could be applied to PBMC from a metastatic rectal cancer patient.
MATERIALS AND METHODS
Cell culture
The melanoma cell line A375 and the CRC cell lines LoVo, RKO, HCT116, HT29, SW403, SW620, SW837, SW480, Colo205, DLD1, and LS174T were obtained from the American Type Culture Collection (Manassas, VA). The colonic adenoma-derived cell lines AA/C1, RG/C2, and LT97 were kindly provided by Dr. William Grady of FHCRC. A375 was cultured in Roswell Park Memorial Insititute (RPMI) medium 1640 with 10 mM hydroxyethyl piperazineethanesulfonic acid (HEPES), 1% penicillin-streptomycin, 1% L-glutamine, 1% sodium pyruvate, and 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA). CRC and adenoma lines were cultured in Dulbecco’s Modified Eagle Medium (DMEM) with 1% PS and 10% FBS (Invitrogen). SW480 was transduced with the firefly luciferase gene using retrovirus that was generated by transfecting Phoenix Ampho packaging cells (National Gene Vector Biorepository, Indianapolis, IN) with MSCV-Luciferase PGK Hygro (Addgene plasmid 18782 from Dr. Scott Lowe, Cold Spring Harbor Laboratory). Transfected cells were subsequently selected with hygromycin 200 μg/mL (Sigma-Aldrich, St. Louis, MO) to produce SW480L, which stably expresses firefly luciferase. Dermal fibroblasts were cultured from skin biopsies as described previously,29 and colon fibroblasts and stromal fibroblasts from a colon adenocarcinoma tumor were cultured from surgical tissue obtained from the Cooperative Human Tissue Network. Fibroblasts were maintained in Dulbecco’s Modified Eagle Medium (DMEM) medium as above.
All PBMC and T cell clones were cultured in RPMI 1640 medium supplemented with 1% penicillin-streptomycin, 10 mM L-glutamine, 50 μM 2-mercaptoethanol (Sigma), and 10% heat-inactivated human serum. T cells were expanded with anti-CD3 antibody (Centocor Ortho Biotech, Horsham, PA) and interleukin-2 (IL-2, Novartis, Basel, Switzerland) as previously described for 13-15 days prior to their use in functional assays.30
Analysis of C-T gene expression
Total RNA from testis (US Biological, Marblehead, MA) and total RNA extracted from cell lines and tissues with AllPrep or RNeasy Plus kit (QIAGEN, Valencia, CA) were converted to cDNA with the First Strand cDNA Synthesis kit (Roche, Indianapolis, IN) using oligo(dT) and random hexamer primers.
A custom RT2 Profiler PCR array (QIAGEN) was designed to permit simultaneous assessment of the expression of genes: BAGE, NY-ESO-1, LAGE-1, CTCFL, DDX53, GAGE1-6, MAGEA1, 2, 3 or 6, 4, 9, 10, and 12, MAGEC2, RAGE, SSX1, 2, and 4, tumor antigen 1 (TAG-1), and PRAME; and human leukocyte antigen-A (HLA-A), β-2-microglobulin (B2M), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Shared primers for MAGEA3 and 6 and for GAGE1 through 6 were used. The relative expression of each gene was determined using the ΔΔCt method, with GAPDH serving as the reference gene and A375 untreated cells as the control group.31 Values were then converted into a visual heatmap using Heatmap Builder 1.1 (a gift of Dr. Euan Ashley, Stanford University).32 Expression of NY-ESO-1 was separately evaluated by real-time quantitative PCR (qPCR), using forward and reverse primer pairs 5′-TGCTTGAGTTCTACCTGCCA-3 ′ and 5 ′-TATGTTGCCGGACACAGTGAA-3′.33 To normalize NY-ESO-1 expression values across different cell types, expression of the GAPDH gene was also evaluated with forward primer 5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse primer 5′-GAAGATGGTGATGGGATTTC-3′.34 The standard curve method was used to calculate NY-ESO-1 expression levels in untreated and DAC-treated cells; the expression of NY-ESO-1 relative in untreated A375 cells was arbitrarily defined as 1.
Western blotting was performed using E978 murine a nti-NY-ESO-1 monoclonal antibody35 (mAb) (Sigma) at 2 μg/mL and a murine anti-human-β-actin mAb (Genscript, Piscataway, NJ) at 0.5 μg/mL, followed by incubation with horseradish peroxidase (HRP)-conjugated goat-anti-mouse IgG at 0.5 μg/mL (Santa Cruz Biotechnology, Santa Cruz, CA). Blots were developed according to the manufacturer’s instructions with the Immobilon Western Chemiluminescent HRP Substrate Kit (Millipore, Billerica, MA).
Immunofluorescence staining
Cells were plated on glass coverslips and treated with 1 μM DAC or medium alone. On day 5, cells were fixed in 3% paraformaldehyde (Sigma) in phosphate buffered saline (PBS) (Invitrogen) and permeabilized with 0.2% Triton X-100 (Sigma) in PBS followed by incubation with 0.5 mg/mL primary E978 antibody 1:200 (Invitrogen) and 2 mg/mL secondary Alexa 594-conjugated goat-anti-mouse antibody 1:500 (Invitrogen) in PBS with 0.1% Tween-20 (Sigma) and 1% bovine serum albumin (Sigma). 4′,6-diamidino-2-phenylindole (DAPI) dihydrochloride (Sigma) was used to visualize nuclei. The coverslips were mounted on a glass slide with Vectashield (Vector Labs, Burlingame, CA) and viewed under fluorescence microscopy.
DAC treatment
In vitro DAC treatment was based on a previously described schedule.21 Briefly, cells were cultured in media with 0.1% Hank’s balanced salt solution (HBSS) (Invitrogen) or 1 μM DAC (Sigma-Aldrich) in HBSS, with fresh medium every 12 hours, for a total of 48 hours, then washed in HBSS and cultured in their respective growth media for an additional 48 hours before being harvested for molecular and functional analyses.
NOD/Scid/IL-2 receptor-γ null (NSG) mice underwent implantation of 5×104 SW480L cells under the right kidney capsule in 30 μL Matrigel (BD Biosciences, Franklin Lakes, NJ) using methods previously described.36 Beginning on day 53 after tumor implantation, they were given intraperitoneal injections HBSS or DAC daily for 5 days and then sacrificed 72 hours after last injection. Tumors were harvested for total RNA, as described.
Generation of endogenous and engineered NY-ESO-1 specific TCR T cells
CD8+ T cells specific for the HLA-A*0201-restricted NY-ESO-1157-165 epitope (SLLMWITQC) were generated by repeatedly stimulating polyclonal CD8+ T cells obtained from a HLA-A*0201+ patient with a NY-ESO-1-expressing synovial sarcoma with irradiated, autologous dendritic cells pulsed with the NY-ESO-1157-165 peptide, then cloning the resulting T cell line by limiting dilution, as previously described.11, 37-40 Cloned CD8+ CTL specific for the HLA-A*0201-restricted minor histocompatibility antigen (mHA) CIPPDSLLFPA encoded by C19Orf48 were isolated by similar technique.29
Under informed consent, blood was collected from a metastatic rectal cancer patient under a clinical protocol approved by the Institutional Review Board of the FHCRC. PBMC were obtained with standard Ficoll-Hypaque technique. The patient was confirmed to be HLA-A*0201+ using the A locus UniTray SSP (Invitrogen). TCR gene transfer into PBMC was performed using a retroviral vector with MSGV1 backbone and encoding the α and β chains of the high-affinity 1G4 TCR specific for NY-ESO-1157-165/HLA-A*0201 and two higher affinity variants of 1G4 with dual amino acid substitutions at positions 95-96 in the α chain (α95:LY) and positions 51-52 in the β chain (β51:AI) packaged with the Phoenix Ampho cell line.28 After activation with CD3/CD28 beads (Dynal) and IL-2, transduced cells were incubated with a fluorochrome-labeled NY-ESO-1157-165/HLA-A*0201 tetramer and antibodies for CD8 and CD4 (BD Biosciences) followed by fluorescence-activated cell sorting (FACS) for CD4+/tetramer+ and CD8+/tetramer+ populations.
Chromium release cytotoxicity assay (CRA)
Target cells were labeled with 51-chromium (51Cr) (Perkin Elmer, Waltham, MA) at 37 °C for 2 hours, then washed and incubated with CTL at varying effector to target (E:T) ratios in triplicate wells in 200 μL of assay medium, which is RPMI 1640 medium supplemented with 1% penicillin-streptomycin, 10 mM L-glutamine, and 10% FBS (Invitrogen), with 25 U/mL IL-2 (Novartis, Basel, Switzerland). Supernatant was collected after 4 hours after incubation of CTL with targets for assessment of radioactivity in a gamma counter (Perkin Elmer).
In vivo bioluminescence imaging of DAC and/or NY-ESO-1-specific CTL clone-treated SW480L
SW480L cells were treated with DAC or HBSS as described above, and on day 5 were plated at 2×104 per well in a 96-well plate with either media or 2×104 native anti-NY-ESO-1 CTL. After overnight incubation, contents of the well were collected, trypsinized, and washed with PBS. All content of the well was then concentrated into 20 μL of growth media and mixed with 30 μL of Matrigel (BD Biosciences) in a 1 mL insulin syringe. NSG mice were irradiated with 250 cGy from a 137Cs source (JL Shepherd Mark I), and cells were surgically implanted under the right kidney capsule. Starting at 2 weeks post-implantation, mice were injected with 40 mg/kg D-luciferin (Caliper Life Sciences, Hopkinton, MA) and imaged on a Xenogen in vivo imaging system (Caliper Life Sciences). Mice were sacrificed when they had lost weight and/or were lethargic. Mouse images were analyzed using Living Image 3.2 software (Caliper Life Sciences).
Real-time Cell Electrical Sensing Assays
Target cells were seeded onto 96-well E-plates (Roche) in duplicate wells numbering 5 × 103 for A375 and dermal fibroblasts and 3 × 104 for SW480 and DLD1 in 100 μL of their respective growth media. After attachment and cell growth was monitored on the real time cell electrical sensing (RT-CES) xCELLigence system (Roche) over 24 hours, 50 μL of growth medium were removed, and replaced with 100 μL of assay medium. Cells were monitored again for another 1-2 hours until the Cell Index stabilized. Then, 50 μL T cell assay medium, effector T cells at a 1:1 ratio to original cell seeding number in assay medium, or 1% Triton X-100 in assay medium were added. Cells were monitored for another 24 hours on the xCELLigence system.
RESULTS
DAC selectively induces expression of NY-ESO-1 and other C-T genes in CRC cells
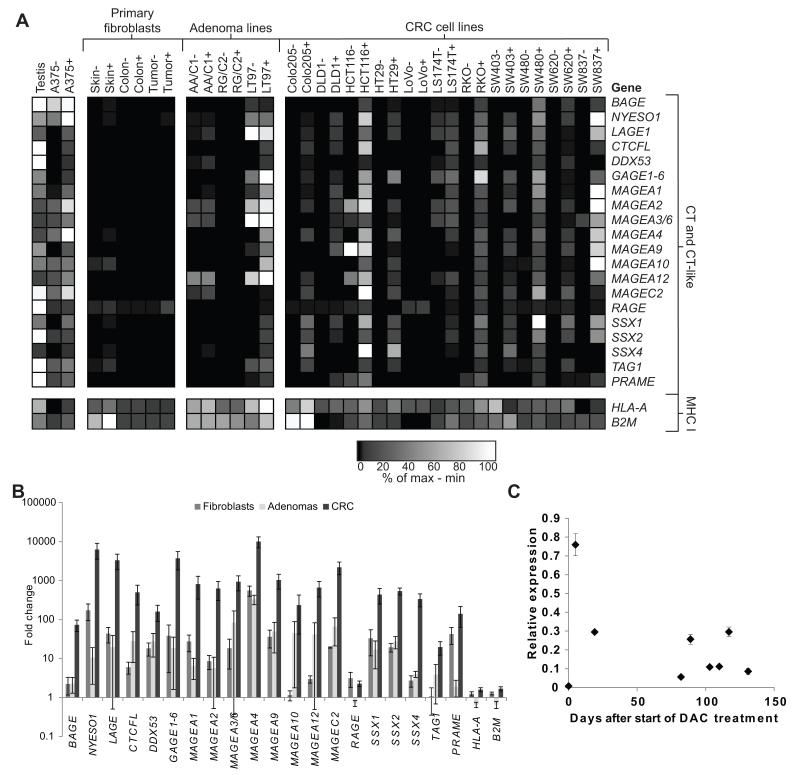
The mRNA expression of C-T genes encoding known T cell epitopes and the major histocompatibility complex (MHC)-encoded genes HLA-A and B2M was evaluated in untreated and DAC-treated CRC cells, colon adenoma lines, and primary fibroblasts.37-47 DAC treatment of CRC cell lines induced expression of most of the C-T genes in a majority of CRC cell lines. (Fig. 1A). In several CRC lines, expression of some C-T genes exceeded levels seen in either testis lysate or the melanoma cell line A375. Although significant increases in expression were observed after DAC treatment in fibroblasts, adenomas, and CRC (Fig. 1B), the overall expression levels of C-T genes in DAC-treated primary fibroblasts was relatively low compared to the levels observed in CRC or colonic adenoma cell lines (Fig. 1A). Two of three adenoma cell lines had detectable C-T gene transcripts at baseline, in the absence of DAC treatment, and the adenoma line LT97 showed induction by DAC of expression of several C-T genes not detected at baseline. There was no significant nor consistent change in expression of either B2M or HLA-A across all cells with DAC treatment. Although peak expression of NY-ESO-1 occurred within the first week of DAC treatment, NY-ESO-1 transcript was detectable in DAC-treated cells up to 130 days at low levels (Fig. 1C).
Figure 1.
Decitabine selectively induces expression of C-T genes in CRC cells. (A) Heat map demonstrating the relative expression levels of selected C-T genes, HLA-A, and B2M in primary dermal fibroblasts, cell lines derived from colonic adenomas, and cell lines derived from colorectal carcinomas, after 48-hour culture in control medium (−) or medium containing 1 μM DAC (+), as determined by RT-qPCR using a custom RT2 Profiler PCR array. GAPDH was used as the reference gene. Data are normalized across each row (corresponding to individual genes), with black representing the lowest expression level and white representing the highest expression level, and the scale indicates the percentile expression level between the minimum and maximum observed values for each gene. (B) Mean fold difference in expression of the indicated genes between control and DAC-treated fibroblasts, colonic adenoma cell lines, and CRC cell lines. (C) SW480 was treated with DAC for 48 hours and continuously cultured in vitro. Cells were collected at separate timepoints post-DAC treatment and assessed for NY-ESO-1 expression by RT-qPCR. GAPDH was used as the reference gene, and 1 represents level of expression in melanoma line A375. Error bars indicate standard error of the mean (SEM).
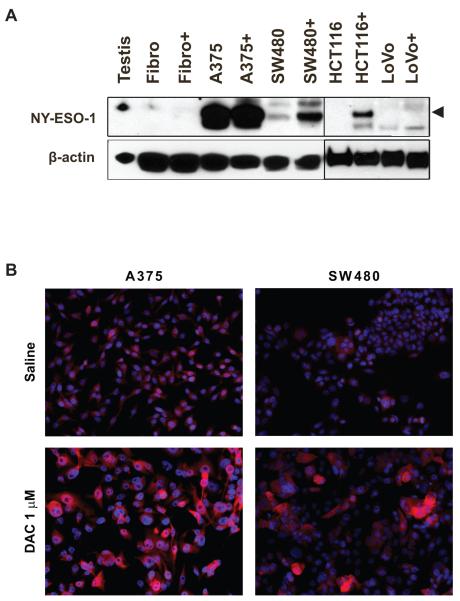
Western blot analysis revealed that DAC induction of NY-ESO-1 expression also correlated with an increase in NY-ESO-1 protein expression (Fig. 2A) in several CRC lines. There was no detectable NY-ESO-1 protein in fibroblasts despite the detection of low levels of mRNA transcript (Fig. 1A). To determine whether the induction by DAC of NY-ESO-1 expression in CRC cells was homogeneous or heterogeneous, immunofluorescence microscopy was performed. Untreated SW480 cells showed little staining with anti-NY-ESO-1 antibody, but DAC-treated SW480 cells showed increased staining in most cells, but the staining was heterogeneous, with higher expression in larger cells (Fig. 2B).
Figure 2.
Decitabine selectively induces expression of NY-ESO-1 protein in CRC cells. (A) Western blot analysis of NY-ESO-1 expression in normal human testis, control or DAC-treated dermal fibroblasts, and control or DAC-treated CRC cell lines. Levels of β-actin were assessed to confirm equal protein loading between paired samples. Arrowhead indicates position of NY-ESO-1 band. (B) Immunofluorescence analysis of NY-ESO-1 expression (red) in control or DAC-treated A375 melanoma and SW480 CRC cells. 4′,6-diamidino-2-phenylindole (DAPI) staining (blue) for nuclear visualization was performed, and the images were merged. Magnification is the same in all 4 panels.
In vivo DAC treatment induces C-T gene expression in CRC xenografts
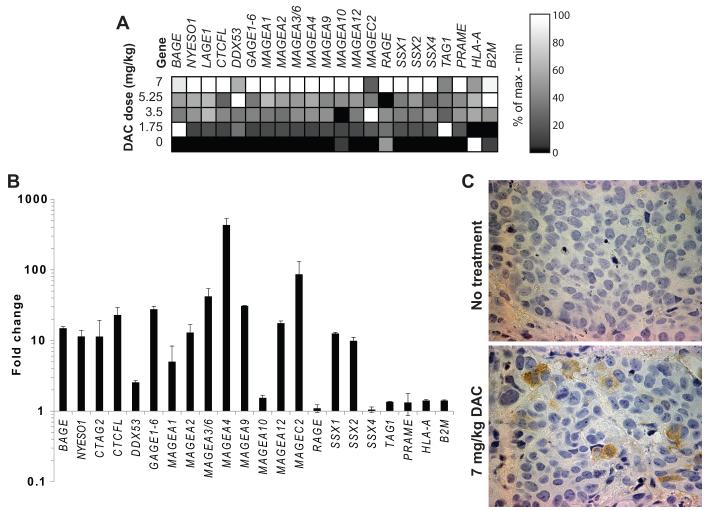
To explore the effects of in vivo DAC treatment, NSG mice bearing SW480L xenografts were injected intraperitoneally with up to 7 mg/kg DAC daily for 5 days, which is equivalent to the standard dose of 20 mg/m2 for 5 days administered to humans with myelodysplastic syndrome (MDS).48 Expression of most C-T genes including NY-ESO-1 increased in a dose-dependent manner (Fig. 3A), with no plateau in expression at the maximum dose. Fold change in C-T gene expression was comparable to that observed with in vitro DAC treatment of CRC lines (Figs. 1B, 3B). NY-ESO-1 protein expression was detectable in a subset of cells within DAC-treated SW480L xenografts by immunohistochemistry (Fig. 3C). Results were consistent with the heterogeneous expression seen with immunofluorescence in vitro (Fig. 2B).
Figure 3.
Decitabine induces expression of C-T genes in CRC xenografts. (A) Heat map demonstrating the relative expression levels of selected C-T genes, HLA-A, and B2M in CRC xenografts after in vivo DAC exposure. NSG mice with established SW480L xenografts were injected IP with 0, 1.75, 3.5, 5.25, or 7 mg/kg DAC daily for 5 days, then sacrificed for tumor harvest and RNA extraction 48 hours later. GAPDH was used as the reference gene. Data are normalized across each gene (in columns), with black representing the lowest expression level and white representing the highest expression level, and the scale indicates the percentile expression level between minimum and maximum observed values for each gene. (B) Fold difference in expression of selected C-T genes, HLA-A, and B2M, between xenografts from control and DAC-treated mice. Data are mean values +/− SEM. (C) Immunohistochemical staining for NY-ESO-1 in SW480L xenografts from mice treated with saline or 7 mg/kg DAC given IP daily for 5 days.
DAC selectively sensitizes CRC cells to recognition by NY-ESO-1-specific CTL
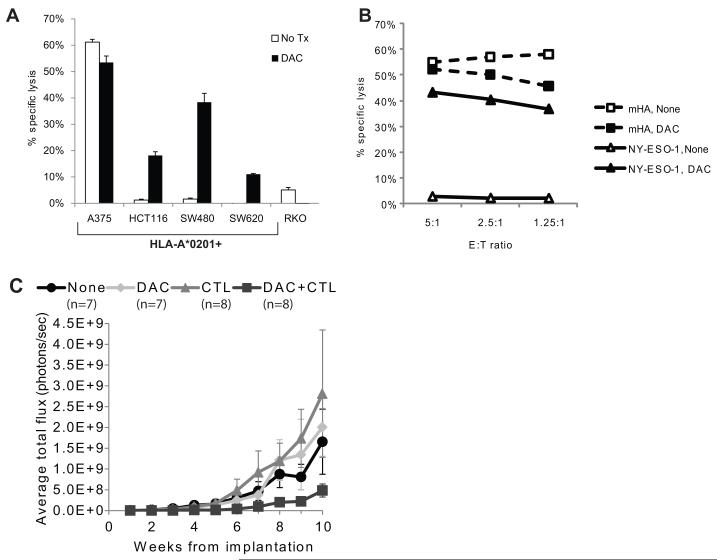
A CD8+ HLA-A*0201-restricted CTL clone specific for NY-ESO-1157-165, derived from a patient with a NY-ESO-1+ synovial sarcoma, was used to determine whether DAC-induced expression of NY-ESO-1 transcript and protein correlates with enhanced recognition by NY-ESO-1-specific CTL. A panel of HLA-A*0201+ (HCT116, SW480, and SW620) and HLA-A*0201− (RKO) CRC cells were treated with DAC and tested for recognition by NY-ESO-1157-165-specific CTL. Recognition of SW480, HCT116, and SW620 in a 4-hour cytotoxicity assay was enhanced by DAC treatment (Fig. 4A). As expected, RKO was not recognized. DAC-treated fibroblasts were also not recognized (data not shown). Adenoma lines were not tested for CTL recognition, since none of the 3 lines used for our studies were HLA-A*0201+.
Figure 4.
CD8+ NY-ESO-1-specific T cells selectively recognize DAC- but not sham-treated CRC cells. (A) Recognition of 51Cr-labeled sham- or DAC-treated CRC cell lines by a HLA-A*0201-restricted, NY-ESO-1157-165-specific T cell clone, isolated from a patient with a NY-ESO-1+ synovial sarcoma, in a 4- hour cytotoxicity assay at an effector:target (E:T) ratio of 5:1. Data are mean values +/− SEM. (B) Recognition of sham-treated (open symbols) or DAC-treated (filled symbols) HLA-A*0201+ SW480 CRC cells by CD8+ HLA-A*0201-restricted CTL specific for NY-ESO-1157-165 (triangles) or the HLA-A*0201-restricted minor histocompatibility antigen encoded by C19orf48 (squares).29 (C) Serial in vivo bioluminescence imaging of CRC xenografts in NSG mice established from SW480L cells that were incubated in vitro overnight in CTL medium alone, medium supplemented with DAC at 1 μM, medium containing NY-ESO-1157-165-specific CTL at a 1:1 ratio, or medium with 1 μM DAC and NY-ESO-1157-165-specific CTL at a 1:1 ratio, then implanted under the kidney capsule. Mice were imaged weekly on a Xenogen in vivo imaging system after injection of D-luciferin. Shown are average total fluxes in each treatment group as a function of the number of days after implantation of SW480L cells. Data are mean values +/− SEM.
To explore whether DAC treatment non-specifically improved T cell recognition of SW480, we compared the cytolytic activity of a CD8+ HLA-A*0201-restricted T cell clone specific for a minor histocompatibility antigen (mHA) that is encoded by C19orf48 and expressed in SW480 cells29 and the NY-ESO-1157-165-specific CTL clone against DAC-treated SW480 cells (Fig. 4B). No significant difference in recognition by the mHA-specific CTL was seen between control and DAC-treated cells, nor did DAC induce expression of C19orf48 (data not shown). Thus, DAC treatment does not globally enhance killing of SW480 by all HLA-A*0201-restricted CTL.
To explore whether treatment with DAC and NY-ESO-1-specific T cells could inhibit engraftment or growth of CRC xenografts in immune-deficient mice, SW480L cells were cultured overnight in (i) control medium, (ii) medium supplemented with 1 μM DAC, (iii) medium containing NY-ESO-1157-165-specific CTL at a T cell:SW480L ratio of 1:1, or (iv) medium supplemented with 1 μM DAC and containing NY-ESO-1157-165-specific CTL at a T cell:SW480L ratio of 1:1, and then implanted under the kidney capsule of sublethally irradiated NSG mice. Mice were imaged weekly on a Xenogen in vivo imaging system after injection of D-luciferin. Growth of tumors as measured by luminescence was observed in all 4 groups of mice (no treatment, DAC, CTL, and DAC+CTL) (Fig. 4C). Although a 1:1 effector to target (E:T) ratio was not sufficient to completely prevent engraftment, growth of xenografts derived from DAC+CTL treated cells was slower than in the other 3 treatment groups. Two-tailed P-values of DAC+CTL compared to other groups using Mann-Whitney U test were significant for no treatment (p = 0.049) and CTL groups (p = 0.049) and trended towards significance for the DAC group (p = 0.096).
NY-ESO-1 specific T cells can be generated from PBMC of a CRC patient
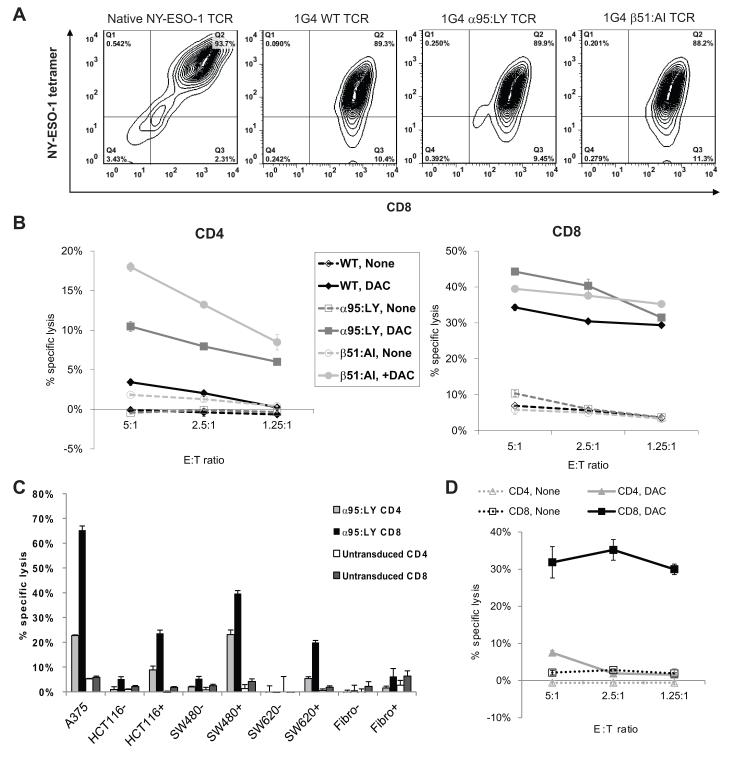
Genes encoding the α and β chains of a TCR, termed 1G4, specific for the HLA-A*0201/NY-ESO-1157-165 epitope, as well as variants (termed α95:LY and β51:AI) of this TCR with dual amino acid substitutions in the α and β chains, respectively, that show enhanced affinity for this epitope,28 were introduced via retroviral transduction into PBMC collected from a patient with metastatic rectal cancer who had not yet received any chemotherapeutic treatment. Transduced PBMC were sorted on the basis of HLA-A*0201/NY-ESO-1157-165 tetramer binding as well as CD4 and CD8 expression, and CD4+tet+ and CD8+tet+ populations were expanded for further study. Transduced T cells were identified by staining with a mAb specific for Vβ 13.1, the protein product of the TCRβ variable (V) gene segment used in the 1G4 TCR, and the HLA-A*0201/NY-ESO-1157-165 tetramer (Fig. 5A).
Figure 5.
Polyclonal CD8+ and CD4+ T cells from a HLA-A*0201+ patient with metastatic rectal cancer transduced with retroviruses encoding the α and β chains of a HLA-A*0201-restricted, NY-ESO-1157-165-specific TCR recognize DAC-treated HLA-A*0201+ CRC cells. PBMC collected from the patient prior to the receipt of adjuvant chemotherapy were transduced with retroviruses encoding the wild type 1G4 NY-ESO-1157-165-specific TCR or its α95:LY or β51:AI variants, then analyzed by flow cytometry with a NY-ESO-1157-165/HLA-A*0201 tetramer and anti-CD4 or anti-CD8 mAbs, and tested for cytolytic activity against CRC targets. (A) Flow cytometric analysis of a natively CD8+ NY-ESO-1157-165-specific CTL clone and polyclonal CD8+ T cells from the CRC patient transduced with retroviruses encoding the wild type or variant 1G4 TCRs. (B) Cytolytic activity of patient T cells transduced with the wild type 1G4 TCR (black) or its α95:LY (dark gray) or β51:AI (light gray) variants against saline- or DAC-treated SW480 cells. (C) Cytolytic activity of CD4+ (light gray) or CD8+ (black) 1G4 α95:LY TCR-transduced patient T cells against control (−) or DAC-treated (+) HLA-A*0201 dermal fibroblasts and CRC cells at E:T 5:1. (D) Cytolytic activity of polyclonal CD4+ (gray) or CD8+ (black) T cells that were isolated from the same rectal cancer patient after 7 months of adjuvant chemotherapy, then transduced with the 1G4 α95:LY variant TCR, against control (broken lines) or DAC-treated (solid lines) SW480 cells. Data shown are mean values +/− SEM.
SW480 cells treated with DAC were efficiently recognized by CD8+ cells transduced with all 3 of the 1G4 TCR variants and by CD4+ cells transduced with the α95:LY and β51:AI variants – similar to what has been observed with melanoma cells (Fig. 5B).28 Since adoptive therapy with polyclonal autologous T cells transduced the α95:LY variant TCR has shown effectiveness in clinical trials in patients with NY-ESO-1-expressing tumors,15 we chose to study its activity against DAC-treated CRC lines in greater depth. At a 5:1 E:T ratio, the 1G4 α95:LY-transduced CD4+ and CD8+ cells recognized DAC-treated HLA-A*0201+ CRC lines but not DAC-treated fibroblasts in a 4-hour cytotoxicity assay. Furthermore, the 1G4 α95:LY-transduced CD8+ T cells exhibited equal avidity for DAC-treated HLA-A*0201+ CRC lines as the natively HLA-A*0201-restricted, NY-ESO-1157-165-specific T cells used in earlier experiments (Fig. 5C). No recognition of DAC-treated CRC cells was seen with untransduced CD4+ or CD8+ T cells.
To determine whether treatment with standard cytotoxic chemotherapy would negatively affect the ability to generate TCR-transduced NY-ESO-1157-165-specific T cells, a second blood draw was obtained from the same patient after 3 months of treatment with 5-fluorouracil, oxaliplatin, and irinotecan (FOLFOXIRI), 3 months of capecitabine, irinotecan, and bevacizumab (CAPIRI+B), and 1 month of single agent capecitabine, and PBMC were again transduced with the 1G4 α95:LY TCR. There did not appear to be any significant difference between the recognition of DAC-treated SW480 cells by TCR-transduced T cells generated either before or after the patient’s treatment with chemotherapy (Fig. 5B, 5D).
Dynamic cytotoxicity measurement based on electrochemical impedance
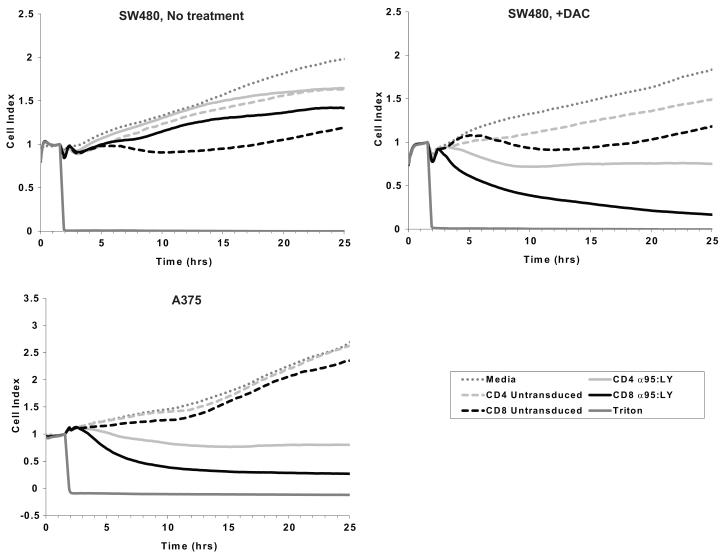
To evaluate the cytolysis of DAC-treated CRC cells by 1G4 TCR-transduced, NY-ESO-1-specific T cells in real time, a RT-CES assay was used. Control or DAC-treated (1 μM for 48 hours) HLA-A*0201+ A375, SW480, DLD1, and dermal fibroblasts were trypsinized, seeded on E-plates 24 hours after last treatment, and allowed to attach and grow for another 24 hours. Control (untransduced) or TCR-transduced T cells were added to the wells at a 1:1 T cell:seeded target cell ratio. At E:T ratios above 1:1, target cell growth was slowed non-specifically (figure, supplemental digital content 1). Sustained drops in impedance, measured as a Cell Index (CI), were observed only when DAC-treated SW480 or natively NY-ESO-1+ A375 cells were co-incubated with 1G4 α95:LY TCR-transduced CD4+ or CD8+ T cells (Fig. 6). No significant decrease in Cell Index was seen when control or DAC-treated DLD1 cells, which do not express surface HLA class I molecules, or DAC-treated HLA-A*0201+ fibroblasts were cultured on the E-plates with either control (untransduced) or TCR-transduced T cells (figure, supplementary digital content 2). A transient but non-sustained decrease in CI was observed when untransduced CD8+ T cells were incubated with either control or DAC-treated SW480 cells. The data from these studies are qualitatively consistent with the observations made with conventional cytotoxicity assays based on 51Cr release (Fig. 5), but provide additional information about the dynamics of cytotoxicity mediated by TCR-transduced T cells. Much of the observed decline in CI occurred after the conventional 4-hour end point of a 51Cr release assay, and was evident at low E:T ratios, suggesting that 1G4 TCR-transduced T cells mediate more potent cytolysis of DAC-treated SW480 cells than the results of 4-hour 51Cr release assays would suggest.
Figure 6.
Dynamic monitoring of NY-ESO-1157-165-specific T cell-mediated cytotoxicity against saline- (upper left panel) or DAC-treated (upper right panel) SW480 CRC cells and control NY-ESO-1+ A375 melanoma cells (lower left panel) using a real time cell electrical sensing system. Cells were seeded in duplicate on E-plates and allowed to attach and grow for 24 hours. Beginning at time 0, the tumor cells were cultured in T cell assay medium for 1.5 hours (dark gray dotted line), after which they were cultured in medium containing CD4+ (dashed light gray line) or CD8+ (dashed black line) untransduced T cells, CD4+ (solid light gray line) or CD8+ (solid black line) 1G4 α95:LY TCR-transduced T cells, or 1% Triton X-100 (solid dark gray line). T cells were added to the E-plates at a T cell:seeded target cell ratio of 1:1. Electrical impedance across the E-plates was continuously monitored every 5 minutes for the next 24 hours, and is reported as a Cell Index, which is normalized to 1 at the time of addition of T cells. Cell indices plotted in the figure represent the mean of duplicate samples.
DISCUSSION
Adoptive transfer of tumor-reactive or tumor-antigen specific T cells has shown objective responses in patients with solid tumors such as melanoma.10-15 The use of adoptive T cell therapy for colorectal cancer, however, has been slowed by the lack of acceptable target antigens. Adoptive transfer of genetically engineered T cells specific for carcinoembryonic antigen (CEA) has led to objective responses in CRC patients, but this was associated with dose limiting toxicity due to inflammatory colitis.49 C-T antigens are restricted in expression to germ cells, which are immunoprivileged, and cancer lines, making them ideal targets for immunotherapy. However, C-T antigens are not commonly expressed in CRC. The results presented here demonstrate that the DNA hypomethylating drug DAC potently and induces expression of NY-ESO-1 and several other C-T genes that are not normally expressed in CRC cells and encode T cell antigens.40 DAC-induced C-T antigens could therefore serve as targets for adoptive T cell therapy in CRC. Shown here for the first time, DAC induction of NY-ESO-1 expression in CRC cells correlates with enhanced recognition by NY-ESO-1157-165-specific CTL. Moreover, although they expressed low levels of NY-ESO-1 transcript, DAC-treated untransformed fibroblasts did not express NY-ESO-1 protein and were not recognized by NY-ESO-1 specific CTL, suggesting that induction of NY-ESO-1 antigen occurs primarily in malignant cells.
In addition to its effects on expression of C-T genes, DAC may act on other pathways in CRC. Several genes playing important roles in the pathogenesis of CRC, particularly tumor suppressor genes, are frequently abnormally methylated in colon tumors.50 Abnormal CpG methylation may be an early event in CRC tumorigenesis and is detected in a majority of tubulovillous and villous adenomas.50, 51 The adenoma lines studied had varying levels of C-T gene transcription and response to DAC (Fig. 1A) compared to the CRC lines, in which the majority of C-T genes were suppressed but inducible by DAC treatment, suggesting that activation of C-T genes occurs at the adenoma state followed by their inactivation by hypermethylation as they progress towards more invasive and proliferative adenocarcinoma. A distinct subtype of CRC is the CpG island methylator phenotype (CIMP), in which a high proportion of genes are hypermethylated. Our studies demonstrate that DAC induces expression of C-T genes in both CRC lines with the CIMP phenotype such as Colo205, DLD1, HCT116, HT29, and RKO as well as lines SW403, SW480, and SW620 that do not have this phenotype.52 Thus, hypermethylation and DAC-mediated demethylation of C-T genes seems to occur independently of CIMP status.
The induction of C-T gene expression by DAC has been shown in a wide spectrum of malignancies with varying histologies, but the extent to which enhanced C-T gene expression enables recognition of tumor cells to immune effectors has not been extensively explored. Sensitization of DAC-treated tumor cells to lysis by NY-ESO-1-specific CTL has been reported in a limited number of other tumor types.20, 21 We have also observed in vitro lysis of DAC-treated breast, ovarian, and kidney cancer and leukemia lines by NY-ESO-1 specific CTL (data not shown). While we were able to detect in vivo expression of NY-ESO-1 in DAC-treated xenografts, the induced expression was overall low but heterogeneous (Fig. 3). As C-T genes have enhanced expression in glioma stem cells versus more differentiated cells,53 DAC-treated CRC cells with higher NY-ESO-1 expression may represent a cancer stem cell population, although further investigation is needed. Also, since there was no plateau of increasing C-T gene expression in response to DAC dose escalation up to the human equivalent for MDS treatment, additional cycles of DAC or increased dosing could improve protein expression levels for T cell recognition.
In order for adoptive T cell therapy against NY-ESO-1 to be feasible in CRC, autologous NY-ESO-1 specific T cells would need to be generated from each CRC patient to be treated. However, isolating NY-ESO-1 specific CTL could prove difficult, given that only 7% of CRC patients have evidence of spontaneous immunity to NY-ESO-1.17 Retroviral transfer of TCR has emerged as a reliable and efficient method to generate autologous antigen-specific T cells from the peripheral lymphocytes of patients. The 1G4 αLY:95 HLA-A*0201/NY-ESO-1157-165 specific TCR has generated objective responses in synovial sarcoma and melanoma patients with NY-ESO-1-expressing tumors.15 In addition to CRC, T cells transduced with NY-ESO-1 specific 1G4 TCR recognize other DAC-treated solid tumor cell lines.24 Although there is a theoretical risk of off-target autoimmune recognition due to mispairing of transduced TCR α and β chains with endogenous TCR α and β chains, there have been no reports of autoimmune toxicity using the 1G4 αLY:95 TCR.15 Also, it is unknown whether DAC-treatment would unmask expression of NY-ESO-1 in normal tissues leading to an autoimmune response from NY-ESO-1 specific T cells, but DAC-treated primary fibroblasts and other non-transformed cell lines such as kidney epithelial cells are not recognized by NY-ESO-1 specific T cells (data not shown).
The 51-chromium release assay has been widely used to assess cytolytic activity of T cells.54 However, there are certain aspects of the assay that limit its usefulness in studying adherent cell lines. Adherent cells are typically placed into suspension for labeling and incubation with T cells thereby removing their attachment to a growth surface, which is not reflective of their normal growth environment. Moreover, the utility of the assay is limited beyond a few hours as 51-chromium is released spontaneously from labeled cells and raises the background activity. As opposed to CRA that provides cytotoxicity data from a single, early time point, RT-CES has been validated as a label-free, non-invasive assay to assess dynamic changes in cytotoxicity from NK cells55, 56 and chemotherapeutic agents.57 These studies suggest for the first time that RT-CES can also be applied to study T-cell mediated cytotoxicity toward adherent cell lines and provides additional information about long-term lysis of targets beyond what CRA provides. Moreover, RT-CES appears to be more sensitive in that T cell recognition of targets is more demonstrable at lower E:T ratios compared to CRA.
In summary, data presented here are evidence that DAC coupled with CTL specific for NY-ESO-1 has in vitro activity against CRC cell lines that are HLA-matched. A combination of epigenetic modulation and immunotherapy targeted against C-T antigens could provide a novel systemic treatment for metastatic CRC.
Supplementary Material
Supplemental Figure 1. (A) Cytolytic activity of untransduced polyclonal CD8+ T cells (gray) or CD8+ 1G4 α95:LY TCR-transduced T cells (black) against control (dotted lines) or 1 μM DAC-treated (solid lines) SW480 CRC cells at the indicated E:T ratios, as measured in a conventional 4-hour 51Cr release assay. Data shown represent the mean of triplicate samples +/− SEM. (B) Dynamic monitoring of CD8+ 1G4 α95:LY TCR-transduced, NY-ESO-1157-165-specific T cell-mediated cytotoxicity against control (left panel) or DAC-treated (right panel) SW480 CRC cells on a real time cell electrical sensing system. Cells were seeded in duplicate on E-plates. After 24 hours to allow for attachment and growth of the seeded target cells the medium bathing the E-plate was changed to T cell assay medium (defined as time 0) and the cells were cultured for an additional 1.5 hours, after which they were cultured in either medium alone (dotted dark gray line) or medium containing CD8+ 1G4 α95:LY TCR-transduced T cells at the indicated T cell:seeded SW480 target cell ratios of 5:1, 2.5:1, 1.25:1, or 0.61:1, or 1% Triton X-100 (solid dark gray line). Electrical impedance across the E-plates was continuously monitored every 5 minutes for the next 24 hours, and is reported as a Cell Index, which is normalized to 1 at the time of addition of T cells. Cell indices plotted in the figure represent the mean of duplicate samples.
Supplemental Figure 2. Dynamic monitoring of 1G4 α95:LY TCR-transduced, NY-ESO-1157-165-specific T cell-mediated cytotoxicity against control (left panel) or DAC-treated (right panel) dermal fibroblasts on a real time cell electrical sensing system. Saline- or DAC-treated fibroblasts were seeded in duplicate on E-plates. After 24 hours to allow for attachment and growth of fibroblasts, the medium bathing the fibroblasts was changed to T cell assay medium (defined as time 0) and the cells were cultured for an additional 1.5 hours. After that point, the cells were cultured in either medium alone (dotted dark gray line), medium containing CD4+ (dashed light gray line) or CD8+ (dashed black line) untransduced T cells, CD4+ (light gray solid line) or CD8+ (black solid line) 1G4 α95:LY TCR-transduced T cells, or 1% Triton X-100 (solid dark gray line). T cells were added to seeded target cells at a ratio of 1:1. Impedance across the E-plates was continuously monitored every 5 minutes for the next 24 hours, and is reported as a Cell Index, which is normalized to 1 at the time of addition of T cells. Cell indices plotted in the figure represent the mean of duplicate samples.
ACKNOWLEDGEMENTS
We thank Cynthia Nourigat, Melissa Comstock, and LaKeisha Perkins of the FHCRC NOD/Scid Core Facility for their assistance with the murine xenograft studies, and Sheila Ojeaburu of FHCRC and Dr. Pierre Del Moral of Roche for their technical assistance. We also thank the patients who have donated their blood and tissues for our work. This work was supported by the Irvington Institute Fellowship Program of the Cancer Research Institute (JC), an ASCO Cancer Foundation Young Investigator Award and an AACR-Colorectal Cancer Coalition Fellows Grant (JC), a Clinical Scientist Award in Translational Research from the Burroughs Wellcome Fund (EHW), the J. Orin Edson Fund for Immunotherapy, and NIH Grants 5 P30 CA015704-34 and 5 P30 DK56465.
Sources of Funding:
AACR and Fight Colorectal Cancer, ASCO, Cancer Research Institute, Burroughs Wellcome Fund, J. Orin Edson Fund, and NIH awards 5 P30 CA015704-34 and 5 P30 DK56465
Footnotes
Financial Disclosure: All authors have declared there are no financial conflicts of interest in regards to this work.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Boyle P, Levin B. World cancer report 2008. IARC Press; Lyon: 2008. [Google Scholar]
- 2.Chiba T, Ohtani H, Mizoi T, et al. Intraepithelial CD8+ T-cell-count becomes a prognostic factor after a longer follow-up period in human colorectal carcinoma: possible association with suppression of micrometastasis. Br J Cancer. 2004;91:1711–7. doi: 10.1038/sj.bjc.6602201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naito Y, Saito K, Shiiba K, et al. CD8+ T cells infiltrated within cancer cell nests as a prognostic factor in human colorectal cancer. Cancer Res. 1998;58:3491–4. [PubMed] [Google Scholar]
- 4.Menon AG, Janssen-van Rhijn CM, Morreau H, et al. Immune system and prognosis in colorectal cancer: a detailed immunohistochemical analysis. Lab Invest. 2004;84:493–501. doi: 10.1038/labinvest.3700055. [DOI] [PubMed] [Google Scholar]
- 5.Phillips SM, Banerjea A, Feakins R, Li SR, Bustin SA, Dorudi S. Tumour-infiltrating lymphocytes in colorectal cancer with microsatellite instability are activated and cytotoxic. Br J Surg. 2004;91:469–75. doi: 10.1002/bjs.4472. [DOI] [PubMed] [Google Scholar]
- 6.Koch M, Beckhove P, Op den Winkel J, et al. Tumor infiltrating T lymphocytes in colorectal cancer: Tumor-selective activation and cytotoxic activity in situ. Ann Surg. 2006;244:986–92. doi: 10.1097/01.sla.0000247058.43243.7b. discussion 992-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galon J, Costes A, Sanchez-Cabo F, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–4. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 8.Pages F, Berger A, Camus M, et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med. 2005;353:2654–66. doi: 10.1056/NEJMoa051424. [DOI] [PubMed] [Google Scholar]
- 9.Mlecnik B, Tosolini M, Kirilovsky A, et al. Histopathologic-based prognostic factors of colorectal cancers are associated with the state of the local immune reaction. J Clin Oncol. 2011;29:610–8. doi: 10.1200/JCO.2010.30.5425. [DOI] [PubMed] [Google Scholar]
- 10.Rosenberg SA, Yannelli JR, Yang JC, et al. Treatment of patients with metastatic melanoma with autologous tumor-infiltrating lymphocytes and interleukin 2. J Natl Cancer Inst. 1994;86:1159–66. doi: 10.1093/jnci/86.15.1159. [DOI] [PubMed] [Google Scholar]
- 11.Yee C, Thompson JA, Byrd D, et al. Adoptive T cell therapy using antigen-specific CD8+ T cell clones for the treatment of patients with metastatic melanoma: in vivo persistence, migration, and antitumor effect of transferred T cells. Proc Natl Acad Sci U S A. 2002;99:16168–73. doi: 10.1073/pnas.242600099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mackensen A, Meidenbauer N, Vogl S, Laumer M, Berger J, Andreesen R. Phase I study of adoptive T-cell therapy using antigen-specific CD8+ T cells for the treatment of patients with metastatic melanoma. J Clin Oncol. 2006;24:5060–9. doi: 10.1200/JCO.2006.07.1100. [DOI] [PubMed] [Google Scholar]
- 13.Dudley ME, Yang JC, Sherry R, et al. Adoptive Cell Therapy for Patients With Metastatic Melanoma: Evaluation of Intensive Myeloablative Chemoradiation Preparative Regimens. J Clin Oncol. 2008;32:5233–5239. doi: 10.1200/JCO.2008.16.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunder NN, Wallen H, Cao J, et al. Treatment of metastatic melanoma with autologous CD4+ T cells against NY-ESO-1. N Engl J Med. 2008;358:2698–703. doi: 10.1056/NEJMoa0800251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robbins PF, Morgan RA, Feldman SA, et al. Tumor Regression in Patients With Metastatic Synovial Cell Sarcoma and Melanoma Using Genetically Engineered Lymphocytes Reactive With NY-ESO-1. J Clin Oncol. 2011;29:917–24. doi: 10.1200/JCO.2010.32.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5:615–25. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 17.Scanlan MJ, Welt S, Gordon CM, et al. Cancer-related serological recognition of human colon cancer: identification of potential diagnostic and immunotherapeutic targets. Cancer Res. 2002;62:4041–7. [PubMed] [Google Scholar]
- 18.Li M, Yuan YH, Han Y, et al. Expression profile of cancer-testis genes in 121 human colorectal cancer tissue and adjacent normal tissue. Clin Cancer Res. 2005;11:1809–14. doi: 10.1158/1078-0432.CCR-04-1365. [DOI] [PubMed] [Google Scholar]
- 19.Gnjatic S, Nishikawa H, Jungbluth AA, et al. NY-ESO-1: review of an immunogenic tumor antigen. Adv Cancer Res. 2006;95:1–30. doi: 10.1016/S0065-230X(06)95001-5. [DOI] [PubMed] [Google Scholar]
- 20.Natsume A, Wakabayashi T, Tsujimura K, et al. The DNA demethylating agent 5-aza-2′-deoxycytidine activates NY-ESO-1 antigenicity in orthotopic human glioma. Int J Cancer. 2008;122:2542–53. doi: 10.1002/ijc.23407. [DOI] [PubMed] [Google Scholar]
- 21.Coral S, Sigalotti L, Altomonte M, et al. 5-aza-2′-deoxycytidine-induced expression of functional cancer testis antigens in human renal cell carcinoma: immunotherapeutic implications. Clin Cancer Res. 2002;8:2690–5. [PubMed] [Google Scholar]
- 22.Weiser TS, Guo ZS, Ohnmacht GA, et al. Sequential 5-Aza-2 deoxycytidine-depsipeptide FR901228 treatment induces apoptosis preferentially in cancer cells and facilitates their recognition by cytolytic T lymphocytes specific for NY-ESO-1. J Immunother. 2001;24:151–61. doi: 10.1097/00002371-200103000-00010. [DOI] [PubMed] [Google Scholar]
- 23.Adair SJ, Hogan KT. Treatment of ovarian cancer cell lines with 5-aza-2′-deoxycytidine upregulates the expression of cancer-testis antigens and class I major histocompatibility complex-encoded molecules. Cancer Immunol Immunother. 2009;58:589–601. doi: 10.1007/s00262-008-0582-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wargo JA, Robbins PF, Li Y, et al. Recognition of NY-ESO-1+ tumor cells by engineered lymphocytes is enhanced by improved vector design and epigenetic modulation of tumor antigen expression. Cancer Immunol Immunother. 2009;58:383–394. doi: 10.1007/s00262-008-0562-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.James SR, Link PA, Karpf AR. Epigenetic regulation of X-linked cancer/germline antigen genes by DNMT1 and DNMT3b. Oncogene. 2006;25:6975–85. doi: 10.1038/sj.onc.1209678. [DOI] [PubMed] [Google Scholar]
- 26.Morgan RA, Dudley ME, Wunderlich JR, et al. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. 2006;314:126–9. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao Y, Zheng Z, Robbins PF, Khong HT, Rosenberg SA, Morgan RA. Primary human lymphocytes transduced with NY-ESO-1 antigen-specific TCR genes recognize and kill diverse human tumor cell lines. J Immunol. 2005;174:4415–23. doi: 10.4049/jimmunol.174.7.4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robbins PF, Li YF, El-Gamil M, et al. Single and dual amino acid substitutions in TCR CDRs can enhance antigen-specific T cell functions. J Immunol. 2008;180:6116–31. doi: 10.4049/jimmunol.180.9.6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tykodi SS, Fujii N, Vigneron N, et al. C19orf48 encodes a minor histocompatibility antigen recognized by CD8+ cytotoxic T cells from renal cell carcinoma patients. Clin Cancer Res. 2008;14:5260–9. doi: 10.1158/1078-0432.CCR-08-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riddell SR, Greenberg PD. The use of anti-CD3 and anti-CD28 monoclonal antibodies to clone and expand human antigen-specific T cells. J Immunol Methods. 1990;128:189–201. doi: 10.1016/0022-1759(90)90210-m. [DOI] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.King JY, Ferrara R, Tabibiazar R, et al. Pathway analysis of coronary atherosclerosis. Physiol Genomics. 2005;23:103–18. doi: 10.1152/physiolgenomics.00101.2005. [DOI] [PubMed] [Google Scholar]
- 33.Vaughan HA, Svobodova S, Macgregor D, et al. Immunohistochemical and molecular analysis of human melanomas for expression of the human cancer-testis antigens NY-ESO-1 and LAGE-1. Clin Cancer Res. 2004;10:8396–404. doi: 10.1158/1078-0432.CCR-04-0809. [DOI] [PubMed] [Google Scholar]
- 34.Barbany G, Hagberg A, Olsson-Stromberg U, Simonsson B, Syvanen AC, Landegren U. Manifold-assisted reverse transcription-PCR with real-time detection for measurement of the BCR-ABL fusion transcript in chronic myeloid leukemia patients. Clin Chem. 2000;46:913–20. [PubMed] [Google Scholar]
- 35.Stockert E, Jager E, Chen YT, et al. A survey of the humoral immune response of cancer patients to a panel of human tumor antigens. J Exp Med. 1998;187:1349–54. doi: 10.1084/jem.187.8.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kreso A, O’Brien CA. Colon cancer stem cells. Curr Protoc Stem Cell Biol. 2008 doi: 10.1002/9780470151808.sc0301s7. Chapter 3:3.1.1-3.1.12. [DOI] [PubMed] [Google Scholar]
- 37.Chen JL, Dunbar PR, Gileadi U, et al. Identification of NY-ESO-1 peptide analogues capable of improved stimulation of tumor-reactive CTL. J Immunol. 2000;165:948–55. doi: 10.4049/jimmunol.165.2.948. [DOI] [PubMed] [Google Scholar]
- 38.Jager E, Chen YT, Drijfhout JW, et al. Simultaneous humoral and cellular immune response against cancer-testis antigen NY-ESO-1: definition of human histocompatibility leukocyte antigen (HLA)-A2-binding peptide epitopes. J Exp Med. 1998;187:265–70. doi: 10.1084/jem.187.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rimoldi D, Rubio-Godoy V, Dutoit V, et al. Efficient simultaneous presentation of NY-ESO-1/LAGE-1 primary and nonprimary open reading frame-derived CTL epitopes in melanoma. J Immunol. 2000;165:7253–61. doi: 10.4049/jimmunol.165.12.7253. [DOI] [PubMed] [Google Scholar]
- 40.Van Der Bruggen P, Zhang Y, Chaux P, et al. Tumor-specific shared antigenic peptides recognized by human T cells. Immunol Rev. 2002;188:51–64. doi: 10.1034/j.1600-065x.2002.18806.x. [DOI] [PubMed] [Google Scholar]
- 41.Shim E, Shim H, Bae J, Lee H, Jeoung D. CAGE displays oncogenic potential and induces cytolytic T lymphocyte activity. Biotechnol Lett. 2006;28:515–22. doi: 10.1007/s10529-006-0008-5. [DOI] [PubMed] [Google Scholar]
- 42.Ayyoub M, Merlo A, Hesdorffer CS, et al. Distinct but overlapping T helper epitopes in the 37-58 region of SSX-2. Clin Immunol. 2005;114:70–8. doi: 10.1016/j.clim.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 43.Adair SJ, Carr TM, Fink MJ, Slingluff CL, Jr., Hogan KT. The TAG family of cancer/testis antigens is widely expressed in a variety of malignancies and gives rise to HLA-A2-restricted epitopes. J Immunother. 2008;31:7–17. doi: 10.1097/CJI.0b013e318159f797. [DOI] [PubMed] [Google Scholar]
- 44.Oehlrich N, Devitt G, Linnebacher M, et al. Generation of RAGE-1 and MAGE-9 peptide-specific cytotoxic T-lymphocyte lines for transfer in patients with renal cell carcinoma. Int J Cancer. 2005;117:256–64. doi: 10.1002/ijc.21200. [DOI] [PubMed] [Google Scholar]
- 45.Godefroy E, Wang Y, Souleimanian NE, et al. Assessment of CD4+ T cells specific for the tumor antigen SSX-1 in cancer-free individuals. Cancer Immunol Immunother. 2007;56:1183–92. doi: 10.1007/s00262-006-0269-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ayyoub M, Stevanovic S, Sahin U, et al. Proteasome-assisted identification of a SSX-2-derived epitope recognized by tumor-reactive CTL infiltrating metastatic melanoma. J Immunol. 2002;168:1717–22. doi: 10.4049/jimmunol.168.4.1717. [DOI] [PubMed] [Google Scholar]
- 47.Ma W, Germeau C, Vigneron N, et al. Two new tumor-specific antigenic peptides encoded by gene MAGE-C2 and presented to cytolytic T lymphocytes by HLA-A2. Int J Cancer. 2004;109:698–702. doi: 10.1002/ijc.20038. [DOI] [PubMed] [Google Scholar]
- 48.Reagan-Shaw S, Nihal M, Ahmad N. Dose translation from animal to human studies revisited. Faseb J. 2008;22:659–61. doi: 10.1096/fj.07-9574LSF. [DOI] [PubMed] [Google Scholar]
- 49.Parkhurst MR, Yang JC, Langan RC, et al. T cells targeting carcinoembryonic antigen can mediate regression of metastatic colorectal cancer but induce severe transient colitis. Mol Ther. 2011;19:620–6. doi: 10.1038/mt.2010.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079–99. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rashid A, Shen L, Morris JS, Issa JP, Hamilton SR. CpG island methylation in colorectal adenomas. Am J Pathol. 2001;159:1129–35. doi: 10.1016/S0002-9440(10)61789-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hinoue T, Weisenberger DJ, Pan F, et al. Analysis of the association between CIMP and BRAF in colorectal cancer by DNA methylation profiling. PLoS One. 2009;4:e8357. doi: 10.1371/journal.pone.0008357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yawata T, Nakai E, Park KC, et al. Enhanced expression of cancer testis antigen genes in glioma stem cells. Mol Carcinog. 2010;49:532–44. doi: 10.1002/mc.20614. [DOI] [PubMed] [Google Scholar]
- 54.Brunner KT, Mauel J, Cerottini JC, Chapuis B. Quantitative assay of the lytic action of immune lymphoid cells on 51-Cr-labelled allogeneic target cells in vitro; inhibition by isoantibody and by drugs. Immunology. 1968;14:181–96. [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu J, Wang X, Xu X, Abassi YA. Dynamic and label-free monitoring of natural killer cell cytotoxic activity using electronic cell sensor arrays. J Immunol Methods. 2006;309:25–33. doi: 10.1016/j.jim.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 56.Glamann J, Hansen AJ. Dynamic detection of natural killer cell-mediated cytotoxicity and cell adhesion by electrical impedance measurements. Assay Drug Dev Technol. 2006;4:555–63. doi: 10.1089/adt.2006.4.555. [DOI] [PubMed] [Google Scholar]
- 57.Solly K, Wang X, Xu X, Strulovici B, Zheng W. Application of real-time cell electronic sensing (RT-CES) technology to cell-based assays. Assay Drug Dev Technol. 2004;2:363–72. doi: 10.1089/adt.2004.2.363. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. (A) Cytolytic activity of untransduced polyclonal CD8+ T cells (gray) or CD8+ 1G4 α95:LY TCR-transduced T cells (black) against control (dotted lines) or 1 μM DAC-treated (solid lines) SW480 CRC cells at the indicated E:T ratios, as measured in a conventional 4-hour 51Cr release assay. Data shown represent the mean of triplicate samples +/− SEM. (B) Dynamic monitoring of CD8+ 1G4 α95:LY TCR-transduced, NY-ESO-1157-165-specific T cell-mediated cytotoxicity against control (left panel) or DAC-treated (right panel) SW480 CRC cells on a real time cell electrical sensing system. Cells were seeded in duplicate on E-plates. After 24 hours to allow for attachment and growth of the seeded target cells the medium bathing the E-plate was changed to T cell assay medium (defined as time 0) and the cells were cultured for an additional 1.5 hours, after which they were cultured in either medium alone (dotted dark gray line) or medium containing CD8+ 1G4 α95:LY TCR-transduced T cells at the indicated T cell:seeded SW480 target cell ratios of 5:1, 2.5:1, 1.25:1, or 0.61:1, or 1% Triton X-100 (solid dark gray line). Electrical impedance across the E-plates was continuously monitored every 5 minutes for the next 24 hours, and is reported as a Cell Index, which is normalized to 1 at the time of addition of T cells. Cell indices plotted in the figure represent the mean of duplicate samples.
Supplemental Figure 2. Dynamic monitoring of 1G4 α95:LY TCR-transduced, NY-ESO-1157-165-specific T cell-mediated cytotoxicity against control (left panel) or DAC-treated (right panel) dermal fibroblasts on a real time cell electrical sensing system. Saline- or DAC-treated fibroblasts were seeded in duplicate on E-plates. After 24 hours to allow for attachment and growth of fibroblasts, the medium bathing the fibroblasts was changed to T cell assay medium (defined as time 0) and the cells were cultured for an additional 1.5 hours. After that point, the cells were cultured in either medium alone (dotted dark gray line), medium containing CD4+ (dashed light gray line) or CD8+ (dashed black line) untransduced T cells, CD4+ (light gray solid line) or CD8+ (black solid line) 1G4 α95:LY TCR-transduced T cells, or 1% Triton X-100 (solid dark gray line). T cells were added to seeded target cells at a ratio of 1:1. Impedance across the E-plates was continuously monitored every 5 minutes for the next 24 hours, and is reported as a Cell Index, which is normalized to 1 at the time of addition of T cells. Cell indices plotted in the figure represent the mean of duplicate samples.