Fig. 1. HLA Class II epitope map of the FVIII C2 domain as predicted by EpiMatrix.
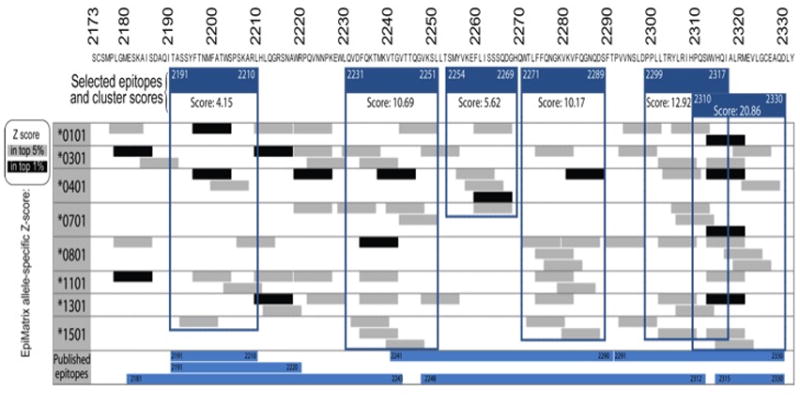
EpiMatrix-predicted 9-mer hits for 8 prevalent HLA class II alleles are aligned along the human FVIII C2 sequence. Any peptide scoring above 1.64 on the EpiMatrix ‘Z’ scale (top 5%) is considered to be a potential epitope (gray bars). Peptides scoring above 2.32 on the scale (top 1%) are extremely likely to bind MHC (black bars). Published epitopes determined by experimental methods are similarly aligned (blue bars). EpiMatrix identified clusters of epitopes spanning residues 2191–2210, 2231–51, 2254–69, 2271–89, 2299–2317 and 2310–30 that are framed in blue. The EpiMatrix algorithm finely maps the positions of published epitopes.
