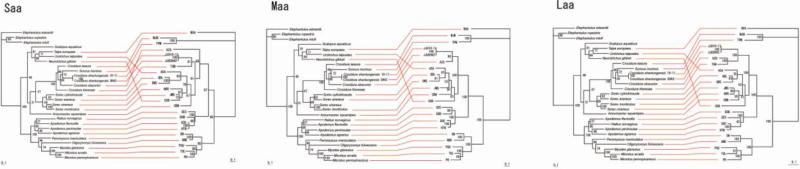
Fig. 3.
Tanglegrams, generated by TreeMap 2.0β, using consensus ML topologies based on the amino acid sequences of the nucleocapsid protein (labeled Saa), Gn and Gc glycoproteins (labeled Maa) and viral RNA-dependent RNA-polymerase (labeled Laa) of JJUV 10–11 and JJUV SH42, and representative rodent-, shrew- and mole-borne hantaviruses and cytochrome b mtDNA sequences of the respective reservoir host species. Node support was derived from 100 ML bootstrap replicates executed on the RAxML BlackBox web server. Virus and host names are provided in the legend to Fig. 2.