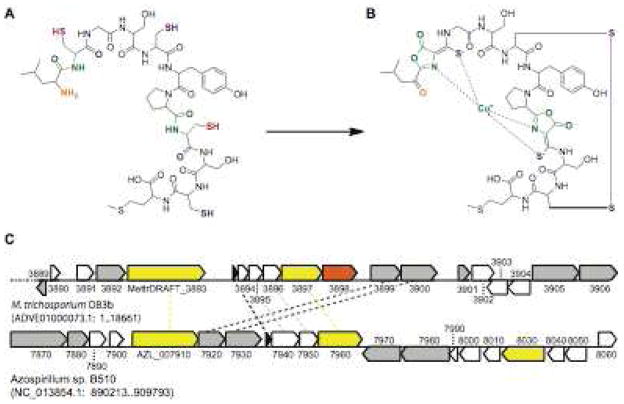
Figure 2.
Possible pathway for biosynthesis of M. trichosporium OB3b Mb. (a) Putative precursor peptide. (b) M. trichosporium OB3b Cu-Mb. Necessary modifications would include replacement of the N-terminal amine group with a carbonyl group (orange), replacement of two cysteines with serines (red) prior to cyclization and formation of the oxazolone rings (green), and replacement of the oxygens in the carbonyl groups adjacent to the two initial cysteines with sulfur to form thioamide groups (blue). A disulfide bond must then be formed (purple) and the resultant apo-Mb loaded with copper (cyan). (c) Genomic neighborhood of the putative precursor gene in M. trichosporium OB3b and Azospirillium sp. B510 genomes. The M. trichosporium OB3b genome has not been fully assembled, and the location of the precursor peptide-containing 18.66 kb contig in relation to the other 172 contigs is not known. The Azospirillum sp. B510 genome is complete; an 18.66 kb region around the precursor peptide is displayed for purposes of comparison. Precursor peptide genes are shown in black, genes with no predicted annotated function are shown in white, genes with predicted annotated functions but unclear relevance to Mb are shown in gray, transporter genes are shown in yellow, and an aminotransferase gene is shown in orange. Potential homologs in the two genomes are connected by dashed lines.