Abstract
DNA microarray analysis of gene expression has become a valuable tool for bioprocessing research aimed at improving therapeutic protein yields. The highly parallel nature of DNA microarray technology allows researchers to assess hundreds of gene simultaneously, essentially enabling genome-wide snapshots. The quality and amount of therapeutic proteins produced by cultured mammalian cells rely heavily on the culture environment. In order to implement beneficial changes to the culture environment, a better understanding of the relationship between the product quality and culture environment must be developed. By analyzing gene expression levels under various environmental conditions, light can be shed on the underlying mechanisms. This paper describes a method for evaluating gene expression changes for cultured NS0 cells, a mouse-derived myeloma cell line, under culture environment conditions, such as ammonia buildup, known to affect product quality. These procedures can be easily adapted to other environmental conditions and any mammalian cell lines cultured in suspension, so long as a sufficient number of gene sequences are publicly available.
Keywords: DNA microarrays, glycosylation, Chinese hamster ovary cells, NS0 cells
1. Introduction
Production of therapeutic proteins in cultured mammalian cells has advanced significantly over the last few decades. Yields during this time have increased from less than ten milligrams per liter to over five grams per liter of product mainly due to improved cell numbers [1]. However, there has not been the same level of improvement in protein quality, specifically with respect to glycosylation and other post-translational modifications that directly impact a therapeutic protein's efficacy [2,3]. Much research has examined how culture variants - including substrate and precursor-feeding, oxygen and carbon dioxide levels, amino acid additions, and osmolarity - influence protein quality [4-22], but currently no robust, reliable means to positively influence protein quality are known [23].
Many studies have investigated gene expression in mammalian cell culture under bioreactor conditions designed to improve protein productivity. Earlier work used real-time quantitative reverse transcription polymerase chain reaction (real-time qRT-PCR) to assess the expression levels of a few particular genes in response to induced culture variants [24-31]. However, these narrow depictions of gene expression did not provide the global analysis necessary to formulate comprehensive models of glycosylation and other post-translational modifications, whereas DNA microarray analysis is beginning to provide adequate levels of detail [32].
DNA microarray technology, with its ability to take genome-wide snapshots, provides a comprehensive tool for gene expression analysis [32] that when combined with proteomic and metabolic data will finally provide the inputs needed to design predictive models for control of post-translational modification, especially glycosylation [23,33,34]. However, DNA microarrays are still most effective for genetically sequenced species, such as mouse cell lines. Thus, the lack of a complete genome prior to July 2011 [35] has limited Chinese hamster ovary (CHO) cell applications, though analysis using private DNA microarrays has yielded insightful results [36-45]. Studies have revealed that genes for vesicle trafficking, endocytosis and cytoskeletal elements critically influenced antibody production [41] and that death receptor- and mitochondria-mediated pathways, not endoplasmic reticulum-mediated pathways, signaled apoptosis [38]. Studies with sodium butyrate have linked its positive effect on protein productivity and improved glycosylation to histone modifications, chaperones, lipid metabolism, and protein processing [39]. When sodium butyrate was combined with lower culture temperatures, genes associated with the Golgi apparatus, cytoskeleton, and signal transduction contributed to the improved protein quality, despite diminished growth rates [42]. Additionally, hyperosmolarity has been investigated in hybridoma cells (a mouse-derived cell line) using DNA microarrays and it has been observed that differential expression of genes associated with catabolism, cell-cycle regulation, apoptosis, regulation of transcription and translation, and transport and signaling pathways play a significant part in cell survival. Surprisingly, hyperosmolarity affected few genes involved in stress-response [43]. The positive affects of copper sulfate on protein productivity and aggregation have been examined via DNA microarrays. The down-regulation of the transferrin receptor and lactate dehydrogenase genes, and the up-regulation of cytochrome P450 family genes were determined to mediate the positive metabolic responses [44]. A weighted gene expression network analysis has been used to identify patterns in gene expression due to relevant bioprocess conditions from 295 DNA microarrays. As expected, genes associated with cell cycle, nucleic acid metabolism, and DNA replication were enriched in the clusters that had positive effects on cell growth and protein production. These clusters may prove useful in identifying biomarkers indicative of high producing CHO cell lines [45]. As these previous results demonstrate, the parallel, high throughput capabilities of DNA microarray technology makes it an ideal tool for evaluating a larger scope of transcriptomic responses to design strategies for improving protein yields and quality.
This article covers step-by-step protocols for mimicking the elevated ammonium stress associated with late-stage bioreactor cultures [46] and using DNA microarrays to analyze the effect of these culture conditions on glycosylation gene expression. While the analysis will focus on NS0 cells (a mouse myeloma cell line used commonly for monoclonal antibody production), these procedures are, in principle, easily adaptable to CHO cells and other relevant suspension cell lines. Additionally, the glycosylation and housekeeping gene sequences that are publicly available could be used with any DNA microarray platform and could be extended to include other post-translational modification genes.
2. Materials
2.1 Cell lines
NS0 cells (ECACC#85110503) from the European Collection of Cell Culture (a mouse myeloma cell line with lymphoblast morphology, non-secreting clone, and cholesterol auxotroph)
(Alternative) Chinese hamster ovary (CHO) cells from the American Tissue Culture Collection (ATCC)
2.2 Cell culture media
ADCF-MAb™ media without L-Glutamine (HyClone) plus L-Glutamine (200 mM) with LS 250 or LS 1000 Lipid Supplement (HyClone), or (Alternative) SyntheChol™ Cholesterol Supplement (Sigma) and Fatty Acid Supplement (Sigma)
(Alternative) EX-CELL® NS0 Serum-Free Medium (Sigma) plus L-Glutamine (200 mM)
(Alternative) CDM4NS0® Medium (Hyclone)
2.3 Cell Culture Reagents, Plasticwear, and Glassware
T-75 (75 cm2) Flasks (e.g. Nunc)
One or two 250 mL Spinner Flask (Kontes Cytostir or similar)
Four or five 1-L Spinner Flasks (Kontes Cytostir or similar)
Orbital Shaker Platform / Multiple Purpose Rotator (e.g. Barnstead Lab-Line)
Stir Plate with at least 4 places for 1-L spinner flasks (e.g. Barnstead/Thermolyne)
5 M NH4Cl stock (in Milli-Q water) (e.g. Sigma)
5 M NaCl stock (in Milli-Q water) (e.g. Sigma)
5 M Proline stock (in Milli-Q water) (e.g. Sigma)
(Alternative/Additional) Computer-controlled fermenter system (e.g. Sartorius BiostatB Plus)
2.4 Cell Counting
Scepter™ 2.0 Handheld Automated Cell Counter (Millipore)
(Alternative) Trypan Blue (e.g. Invitrogen), Microscope (e.g. Olympia), and Hemocytometer
(Alternative) Coulter counter (e.g. Beckman).
2.5 Glucose Analysis
HemoCue® B-Glucose Analyzer (HemoCue)
(Alternative) YSI 2300 STAT Plus™ Glucose & Lactate Analyzer (Yellow Springs Instruments)
(Alternative) NOVA BioProfile® Basic analyzer (NOVA Biomedical)
2.6 RNA Stabilization
Ice bath to quickly cool cells
(Optional) RNAlater® (Ambion)
(Optional) RNAProtect® (Qiagen)
2.7 RNA Isolation and Purification
RNAqueous-Midi® kits (Ambion)
(Alternative) PureYield™ RNA Midiprep System (Promega)
(Alternative) RNeasy® Midi Kit (Qiagen)
2.8 RNA Clean-up (Optional)
RNeasy® Mini Kit (Qiagen)
RiboPure™ Kit (Ambion)
(Optional) RNase-Free DNase Set (Qiagen)
2.9 RNA Quantification
Agilent 2100 Bioanalyzer (Agilent Technologies)
(Alternative) Ribogreen® RNA Quantification kit (e.g. Molecular Probes)
(Alternative) Spectrophotometer with 280 and 260 nm filters (e.g. TECAN)
(Alternative) Gel electrophoresis apparatus (e.g. Nunc)
2.10 DNA Microarray
GLYCOv4 GeneChip® (Affymetrix), not available for commercial use.
(Alternative) Custom Nimblegen DNA microarray
(Altenative) Custom spotted DNA microarray
2.11 Gene Expression Analysis
GeneSpring® (Agilent)
(Alternative) ArrayStar® (DNASTAR)
(Alternative) ORIOGEN v3 (http://www.niehs.nih.gov/research/resources/software/oriogen/index.cfm), not for commercial use.
2.12 Real-time qRT-PCR validation
Access RT-PCR kit (Promega, WI)
SYBR® Green I dye chemistry (Molecular Probes, Eugene, OR)
Primer Express® (Applied Biosystems, Foster City, CA)
Smart Cycler® (Cepheid, Sunnyvale, CA)
3. Methods
3.1 Overview
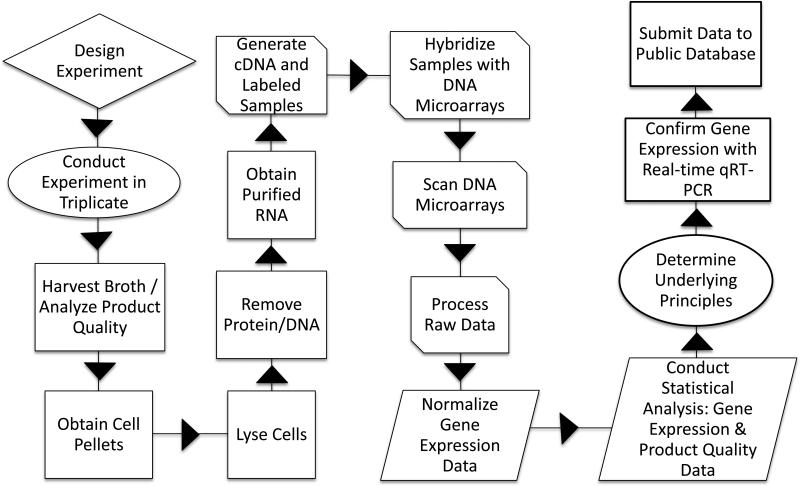
This article will cover the key elements of the procedures needed to assess gene expression changes under bioreactor conditions with particular focus on protein glycosylation and other post-translational modification genes. The factors that should be considered when planning such experiments will be discussed. Although DNA microarrays are central to assessing gene expression levels, much of the experimental procedures described here – including experimental setup and sampling procedures – are independent of the selected DNA microarray platform. Additionally, most DNA microarray scanning software can generate multiple types of specialized files (such as CEL for Affymetrix) as well as tab-delimited text files, and gene expression software programs can read a variety of file types. Due to this flexibility, the experimental procedures mentioned here can be performed fairly independent of the DNA microarray platform to be used. A flow diagram outlining the basic procedure can be found in Figure 1. The details of the experimental design and set up will be discussed in more detail and the reader will be informed of potential options.
Figure 1.
Flow diagram for the collection and analysis of gene expression data. The process begins with the experimental design. Next, the experiments are conducted in triplicate for each condition. From each condition, the samples are harvested and the cells obtained. The product quality can be analyzed from the harvest broth samples. In order to analyze gene expression, the total RNA is separated from the cells and purified. Depending on the DNA microarray platform, the researcher will convert the mRNA to cDNA, or send the total RNA to a DNA microarray contract facility for processing or process in-house. The cDNA is then hybridized with DNA microarray, the DNA microarray is scanned and the data is processed. At this point the processed data will be available for analysis. The processed data is usually imported into a DNA microarray analysis software tool, where the data is normalized. Once the data is normalized, statistical analysis is conducted to identify genes with differential expression between the replicates. Statistical analysis of the product quality is conducted in parallel with the gene expression analysis. A variety of tools can be used to determine underlying gene regulation principles. In order to publish the research results, the data is then submitted to a public database.
3.2 Experimental Design and Set-up
As a means to demonstrate how to set up and run an experiment focused on quantifying expression changes in genes associated with glycosylation and post-translation modification, a step-by-step procedure will be described for a model system. The model system will be NS0 cells exposed to conditions known to affect protein glycosylation, including elevated ammonium [16,46,47] as well as three other conditions. Specifically, the experimental procedure to set up ammonium-stressed, salt-stressed, ammonium-stressed with proline, and control cultures will be described. The salt-stressed will provide a control for osmolarity. The ammonium-stressed with proline will examine the ability of proline to mitigate the negative effects of ammonium [28]. Additionally, a control will normalize volumes, nutrients, and osmolarity. Biological triplicates for each condition will be needed, thus requiring 12 individual culture vessels.
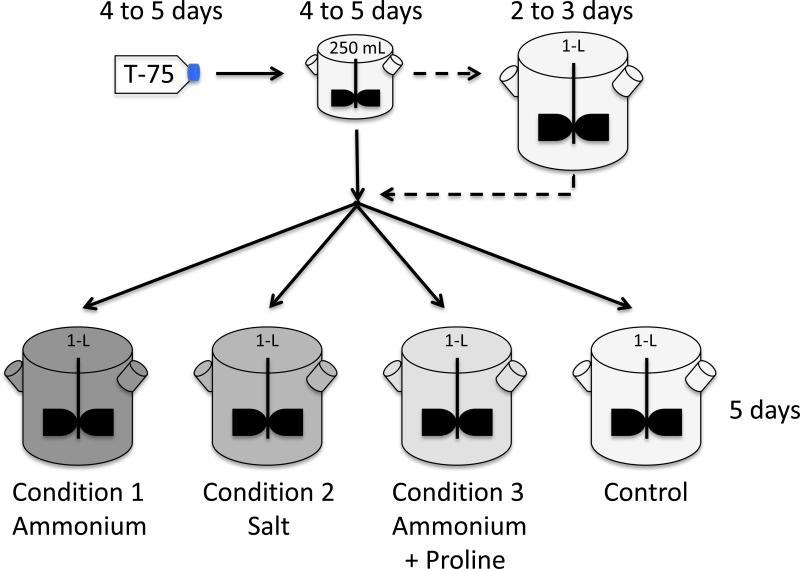
Since the anticipated gene expression changes are likely to be small, relative to the large magnitude changes observed due to a heat-shock, it is important to synchronize the control and “stressed” cultures as much as possible to reduce the growth-associated differences. The use of a common inoculum for the control and stressed conditions synchronizes the culture behavior, which should reduce the noise-to-signal ratio. In Figure 2, a scale-up plan is shown for using a common inoculum to seed four 1-L spinner flasks. These four spinner flasks represent only one-third of the experiment and one complete set of the conditions. This process would need to be repeated two more times to generate the biological triplicates (12 experimental vessels total). Since most CO2 incubators can only hold one multi-place stirrer with four 1-L spinner positions, the procedure outlined in this article runs biological triplicates from three separate inocula, sequentially. It is possible to reduce the overall time by staggering the T-75 flasks and recycling the 250 mL spinner with a one-day delay. This would stagger the four experiment 4-L spinner flasks by one day, allowing sufficient time to harvest, wash, and sterilize the spinner flasks.
Figure 2.
Expansion timeline for NS0 cells to be used in parallel 1-L spinner flasks. NS0 cells are inoculated into a T-75 flask and allowed to expand to approximately 1.5 × 106 cells/mL, which takes 4 to 5 days. The T-75 flask cells (25 mL) are used to inoculate one 250 mL spinner flask with 225 mL fresh media added. If the desired culture volume in the four 1-L spinners is only 500 mL, the cells from one 250 mL spinner flask can be used to inoculate all four 1-L spinners. However, if a higher volume is required in the 1-L spinners, an additional 1-L spinner can be used to expand the seed train. Inoculate the extra 1-L spinner with the entire contents of the 250 mL spinner flask and add up to 750 mL fresh media. Inoculate the control and “stressed” spinner flasks with the same volume of cells from either the 250 mL spinner or the additional 1-L spinner flasks (50 mL to 250 mL depending on desired timeframe).
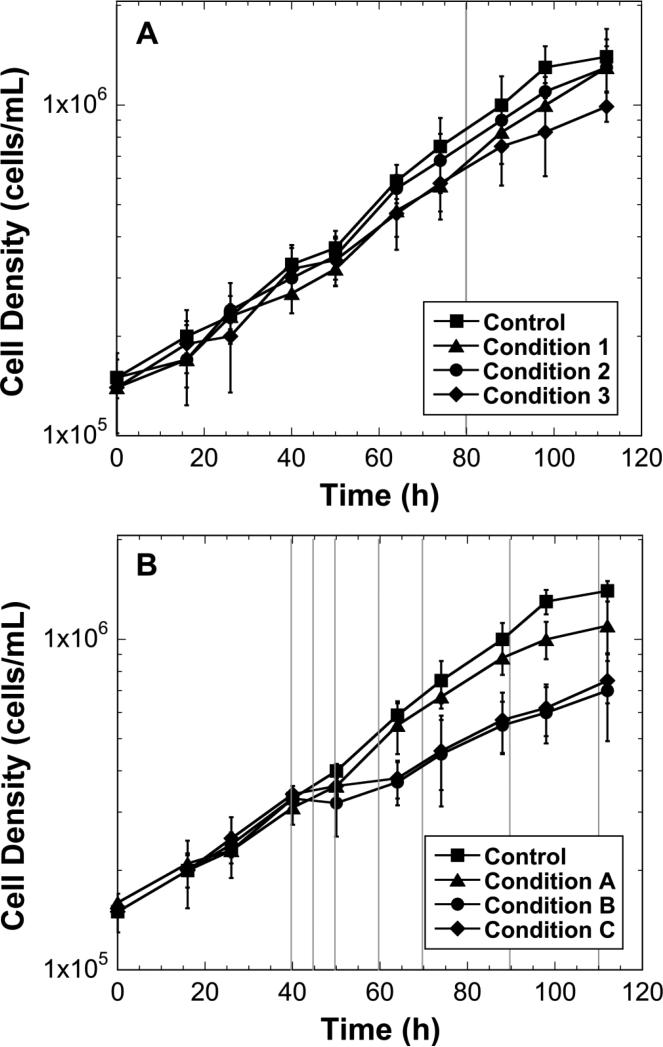
For stresses sufficiently mild as to not alter the growth significantly, inoculating the cells directly into the vessels containing the control and “stressed” media allows for synchronization. An example set of growth profiles are shown in Figure 3A where the stresses were mild, as indicated by the parallel growth profiles. An alternative approach is required if the stresses to be investigated cause significant growth rate changes. In this case, is it best to inoculate all the vessels with cells in control media. This will allow the cells to grow sufficiently to obtain a baseline growth rate and confirm that cultures are not different, and then the stresses can be applied. An example set of growth profiles are shown in Figure 3B that highlights this approach, where the stresses were added at 40 hours. The growth profiles were similar prior to the stress at 40 hours, and divergent after 40 hours.
Figure 3.
Example growth profiles for biological triplicate conditions. A) The conditions being examined did not significantly affect the growth rate; however, the final cell densities were significantly different. In this case, obtaining gene expression analysis data from one time point, late in culture (around 90 hours, indicated by the vertical line) would not have high noise in the data due to a growth effect. B) The conditions being examined significantly affect the growth rate. In this case, the stress was applied at 40 hours. Sampling throughout the culture's growth (before and after the stress) would be desirable, such that gene expression changes due to growth effect could be identified. The total RNA sample times are indicated by the vertical lines.
The sampling frequency and types needed for analysis will directly determine the minimum volumes needed in the experimental vessels. In the model system, 500 mL media was used in 1-L spinner flasks to assure sufficient material throughout the entire experiment. The volume reduction due to sampling should not significantly alter the culture characteristics, such as the mixing dynamics. For the model case, samples would be taken for cell density, cell viability, glucose, lactate, osmolarity, and total RNA. If the cell line of interest expressed a therapeutic recombinant protein, samples analyzing the product quality should be taken periodically. The cell density, cell viability, glucose, and lactate samples would be taken approximately every 12 hours (See Figure 3A). For mammalian cells, the sampling frequency can vary from 10 to 16 hours without missing the culture dynamics; however, it is important that the time intervals used be the same for all replicates such that error bars in time are not needed. In the model case, the sample interval was initially 16 hours, and then alternated between 10 and 14 hours to accommodate more user-friendly work hours (See Figure 3). The volumes withdrawn for these samples should be 5 mL per time point with 10 time points (50 mL total). The frequency of these sample types would be the same for the growth inhibitory stress conditions (See Figure 3B).
For DNA microarrays, a minimum of 10 μg total RNA is usually required. Thus, it is critical that the cell pellet samples for total RNA to be sufficiently large enough to assure sufficient material is obtained. Cultured cells typically have much lower total RNA levels than tissue sample, consequently 1 × 107 cells are a minimum target of cells to assure that 10 μg total RNA is obtained at the necessary concentration (1 μg/μL). For beginning cultures with cell densities in the 2 × 105 cell/mL range, harvesting 50 mL culture media per sample is sufficient. Once the cell densities are greater than 3 × 105 cell/mL, the harvest volumes can be decreased to 15 mL. The recommended RNA isolation kits can handle up to 100 μg total RNA and associated cell debris, so it is unlikely that a kit will be saturated with cell debris. For the non-growth altering stresses, a single, late-stage exponential-phase total RNA sample from each condition would be sufficient to capture gene expression changes due to the stress, which was indicated by the vertical line in Figure 3A. Taken together with the other sample volumes (50 mL), the total anticipated volume change in the vessel is only approximately 65 mL out of 500 mL. For the growth altering stresses, multiple total RNA samples from each condition would be needed to capture gene expression changes due to the stress. For the model experiment shown in Figure 3B as vertical lines, total RNA samples should be taken at 40, 45, 50, 60, 70, 90, and 110 hours to fully capture the potential culture gene expression dynamics. Taken together with the other sample volumes (50 mL), the total anticipated volume change in the vessel would be 155 mL. The similarity of mixing dynamics can be assessed using culture media and tracer dyes at the various anticipated volumes to confirm minimal effects. So, depending on the placement of the stirring paddles, the initial culture volume for the growth altering stressed cultures may need to be increased to between 750 and 1000 mL.
3.3 Culturing NS0 Cells
Standard cell culture techniques should be used to thaw and expand the NS0 cells.
3.3.1 Standard Media Preparation
The ADCF-MAb media requires a cholesterol supplement, since NS0 cells are cholesterol auxotrophs. ADCF-MAb media was designed to be used with the LS 250 lipid supplement (4 mL per liter media; Hyclone) or LS 1000 (1 mL per liter media; HyClone). Alternatively, the cholesterol supplement SyntheChol™ (2 mL per liter, Sigma) and the Fatty Acid Supplement (0.5 mL per liter, Sigma) can be used as well. As needed, add L-glutamine (25 mL of 200 mM L-glutamine stock per liter) to the ADCF-MAb media. If other cell lines or media are used, prepare media appropriately.
The NS0 cells will be inoculated into fresh media at approximately 2 × 105 cells/mL and allowed to reach 1.5 × 106 cells/mL prior to subsequent inoculation. Preparation of the first spinner flask requires a cell density in the T-75 flask greater than 1.0 × 106 cells/mL. For example, 25 mL of 1.0 × 106 cells/mL added to 225 mL of fresh media will result in an initial cell density of 1 × 105 cells/mL in the 250 mL spinner flask. The 250 mL spinner flask will reach approximately 1.0 × 106 cells/mL in about 3 to 4 days. Since the complete ADCF-MAb media supports cell densities of approximately 2 × 106 cells/mL, the cells should be transferred to fresh media in the exponential phase (< 1.5 × 106 cells/mL) to reduce the potential of a growth lag. For NS0 cells, a lead-time of 8 to 12 days is needed for each replicate set. These lead-times can be shortened if the cell line to be examined has faster doubling times than NS0 cells. For example, the higher growth rate of CHO cells compared to NS0 cells results in less time required to expand the cells.
If the phenomenon to be investigated requires high cell density cultures, computer-controlled fermenters operated in a fed-batch manner will be required. The description of fed-batch operation of mammalian cell cultures will not be described; however, there are several research papers that describe fed-batch operation [48-50]. The inoculum for the fermenters will be generated in spinner flasks, as described above. Depending on the number of fermenters available, the conditions to be examine may need to be run sequentially. If sequential operation is required, it is very important to have standardized expansion trains.
3.3.2 Media Preparation
For the model case, three stressed conditions were compared to the control, representing cells cultured in the standard media. In order to minimize osmolarity and nutrient concentration differences, the final volumes in the experimental spinner flasks were kept constant by water additions to the control and other stressed conditions as required (See Table 1). For example, for the ammonium-stressed condition (Condition 1), 2 mL of the 5 M NH4Cl stock was added to the media and 4 mL of Milli-Q water. For the salt-stressed condition (Condition 2), 2 mL of the 5 M NaCl stock was added and 4 mL of Milli-Q water. For the ammonium-stressed plus proline condition (Condition 3), 2 mL of the 5 M NH4Cl stock was added and 4 mL of the 5 M proline stock. For the control culture (Control), 6 mL of Milli-Q water was added. To reduce any potential osmolarity shocks, the salts, proline, ammonia, and water should be added to the medias prior to the addition of cells. If the stresses significantly affect growth after the stress is added, the control culture should have a water addition of equal volume to any chemical addition.
Table 1.
Media additions to induce the stressful condition, while normalizing the nutrient concentrations and osmolality. Amounts listed are per liter of the final media.
| Additions | Control | Condition 1 (10 mM Ammonium) | Condition 2 (10 mM Salt) | Condition 3 (10 mM Ammonium + 20 mM Proline) |
|---|---|---|---|---|
| 5 M NH4Cl | 0 | 2 mL | 0 | 2 mL |
| 5 M NaCl | 0 | 0 | 2 mL | 0 |
| 5 M Proline | 0 | 0 | 0 | 4 mL |
| Milli-Q Water | 6 mL | 4 mL | 4 mL | 0 |
3.3.3 Initial Sampling
Once the vessels are inoculated, the initial cell density should be determined. A Scepter™ 2.0 Handheld Automated Cell Counter or a similar device can be used to obtain the initial cell densities and if cell viability is desired, the Trypan Blue exclusion method can be used. Additionally, samples should be taken for glucose and lactate analysis using a NOVA BioProfile® Basic analyzer or the YSI 2300 STAT Plus™ Glucose & Lactate Analyzer. Procedures for the NOVA BioProfile Basic® analyzer or the YSI 2300 STAT Plus™ Glucose & Lactate Analyzer can be found from the manufacturers.
3.3.4 Sampling
The cultures should be monitored periodically for 110 hours with sampling every 10 to 14 hours. Samples for every time point should minimally include cell density and glucose concentrations. Cell viability and lactate may provide additionally information on cell health. The cell pellets for total RNA samples should be taken as per the experimental design. Protein quality assessment samples should also be taken for systems that express a therapeutic recombinant protein.
3.4 RNA Isolation and Purification
Sampling for total RNA isolation requires some type of stabilization. One method simply rapidly cools the cell broth on ice water (4°C), then centrifuges at 4°C. The cell pellet is then stored at -80°C. Alternatively, a stabilization agent can be added directly to the cell broth for storage. Usually an equal volume of the stabilization agent is needed. Commercially available stabilization agents include RNAlater® Stabilization Solution (Ambion) and RNAprotect® cell reagent (Qiagen). The stabilized cell broth is then either stored at -20° C or -80° C depending on the manufacturer.
Although the RNA extraction kits are designed to inactivate RNAses and other degradative compounds, it is essential that proper sterile practices be used, and that no part of the kits be touched with bare hands, as RNAses are especially abundant on human skin, and could affect reagents and samples. Also, all water and pipette tips used in conjunction with these kits must be nuclease-free. It is best to process all the RNA from all samples together.
3.4.1 RNA Isolation
The RNAqueous® Midi Kit can be used to extract and isolate the total RNA, although many other kits, such as RNeasy® Mini Kits, are also effective. These procedures can be found at http://www.ambion.com and http://www.qiagen.com. Also, the phenol-chloroform extraction method can be utilized. It is sold under the name TRI Reagent (Sigma) and TRIzol® (Invitrogen). Essentially, these procedures all have steps to lyse the cells, remove cellular debris including lipids, proteins and DNA, followed by binding and eluting the total RNA in nuclease-free water.
3.4.2 RNA Purification
Many DNA microarray contract facilities require an additional RNA purification. The RNeasy® Mini Kits can be used for all RNA purifications procedures. Other companies, such as Norgen Biotek offer alternatives; however, one should confirm the compatibility with the DNA microarray contract facility beforehand. Normally, RNA purification kits efficiently remove the majority of DNA without DNAse treatment; however, if any of the mRNA transcripts of interest are in low abundance, an additional DNAse digestion step should be performed. An RNase-Free DNase Set should be used. The procedure for Qiagen's DNAse digestion can be found at www.qiagen.com/hb/rnasefreednaseset. In the model case, the DNAse digestion step was not needed; however, the additional Qiagen RNA clean-up was required by the Consortium for Function Genomics DNA microarray facility.
3.4.3 RNA Quantification
The Ribogreen® RNA Quantification Kit (Molecular Probes) was used to quantify the concentration of all RNA samples, although other alternatives include the Agilent 2100 Bioanalyzer, UV/VIS spectrophotometer, or gel electrophoresis apparatus. The Ribogreen® Quantification Kit procedure can be found at http://hcgs.unh.edu/protocol/realtime/RiboGreen.pdf.
3.5 DNA Microarray
There are several existing DNA microarray technologies; however, for this article, the descriptions will focus on the usage of a high-density oligonucleotide DNA microarray synthesized in situ using photolithography, a method developed by Affymetrix. Other DNA microarray platforms that could be used with the glycosylation and post-translational modification genes include pin-based fluid transfer systems, piezo-based inkjet dispenser systems, and electronic-based addressing systems [51]. Selection of a DNA microarray platform depends on the experimental design, gene sequences available, processing facility availability, and cost. Muyal et al. (2008) provides a good review of the different platform technologies [52]. Further, Larkin et al. (2005) demonstrated that results obtained obtain from Affymetrix and two-color DNA microarray platforms agreed, as long as reliable and consistent methods of identifying genes were used. This makes sense when one considers that the biological and procedural variation exerts a greater effect than platform variation [53].
The specific DNA microarray that will be described is the GLYCOv4 GeneChip® DNA microarrays. The GLYCOv4 GeneChips® are Affymetrix-manufactured DNA microarrays that contain the known glycosylation-related genes for human and mouse. The GLYCOv4 DNA microarrays were constructed for the Consortium for Functional Glycomics with funding from the National Institutes of Health (NIH). The GLYCOv4 GeneChips® are only available through a proposal process to academic researchers from the Consortium for Function Genomics. Alternatively, a researcher could have a custom DNA microarray manufactured containing these glycosylation genes using any platform technology, as all the gene sequences are publicly available.
The GLYCOv4 GeneChip® DNA microarray contains 25 perfect match and mismatch (PM-MM) probes for each of the 1127 mouse glycosylation-related genes and the 119 housekeeping genes. The list of all the included genes and probe sequences can be found at http://www.functionalglycomics.org/static/consortium/resources/resourcecoree.shtml.
The gene types include:
Glycosyltransferases
Glycan degradation proteins
Intercellular protein transport proteins
Nucleotide sugar synthesis and transporter proteins
N-glycan biosynthesis-related proteins
Interleukins and receptors
Growth factors and receptors
Cytokines
Affymetrix DNA microarrays also include numerous control probes on the array. Details of the Affymetrix Eukaryotic Hybridization Controls can be found at http://www.affymetrix.com. Briefly, the GeneChip® Eukaryotic Hybridization Controls contains a mixture of four biotin-labeled cRNA controls with staggered concentrations. The controls provides alignment signals used by the scanning software to provide calibrated intensities that determine “present” and “absent” calls for a gene as well as confirm hybridization and chip integrity.
3.6 RNA Sample Preparation for DNA Microarray
The total RNA samples need to be very concentrated (1 μg/μL) for DNA microarray analysis. Therefore, it may be necessary to precipitate the total RNA after the purification step. For the model case, a vacuum concentrator was used to dry the 10 μg samples, which then were resuspended in 10 μL. Theses samples were sent to the Consortium for Function Genomics. As typical of contract DNA microarray facilities, the total RNA quality and concentration were determined upon arrival with an Agilent 2100 Bioanalyzer. This quality assurance step, allows the contractor to avoid expense of labeling total RNA that has degraded in transit. Alternatively, some contract DNA microarray facilities require the researcher to reverse transcribe the RNA to cDNA, as shipping the more stable cDNA is less problematic. If required to reverse transcribe the RNA on-site, make sure to budget the cost of the reserve transcriptase (~$100 per sample).
At the contract DNA microarray facilities, the RNA will be reverse transcribed in the presence of the fluorescently-labeled dNTP analogues to obtain biotinylated cRNA. The labeled RNA will be hybridized to a DNA microarray, such as the GLYCOv4 GeneChip®, including Affymetrix's GeneChip® Eukaryotic Hybridization Controls. The GLYCOv4 GeneChips ® will be scanned using the Hewlett-Packard GeneArray Scanner G2500A and a processed image file with calculated signal intensities assigned to each oligonucleotide spot on the GLYCOv4 GeneChip® will be created. The data will be reviewed for quality control at the contract DNA microarray facilities. The data files are processed and returned as CEL files (for Affymetrix) to the researcher. Additionally, data regarding the probes is provided. An example of the probe data provide in shown in Table 2 for a GLYCOv4 GeneChip®. Only data for the first 10 probes is shown in Table 2 for brevity. Muyal et al. (2008) provides a good overview description of the DNA microarray process from RNA sample preparation to data acquisition (imaging) [52]. Additionally, the GeneChip® Expression Analysis Technical Manual provides details of the RNA preparation through imaging steps for Affymetrix DNA microarrays.
Table 2.
Example probe set data information for the GLYCOv4 GeneChip®. Information for only the first 10 genes is listed. The GLYCOv4 GeneChip® contains 25 probes per accession number. The accession numbers are sufficient information to create a custom DNA microarray.
| Probeset ID | Category | Sub-category | Common name | Accession Number | Species | Probe Set Score |
|---|---|---|---|---|---|---|
| NM_013630.2_psr1 | CBP: C-Type Lectin | 10-Polycystin | Pkd1 [Polycystin] | NM_013630 | Mouse | 10.6 |
| NM_029686.2_psr1 | CBP: C-Type Lectin | 10-Polycystin | Pkd1l2 [Polycystin 1-like protein 2] | NM_029686 | Mouse | 12.6 |
| NM_181415.3_psr1 | CBP: C-Type Lectin | 11-Attractin | Atrnl1 [Attractin homolog] | NM_181415 | Mouse | 10.5 |
| NM_009730.2_psr1 | CBP: C-Type Lectin | 11-Attractin | Attractin | NM_009730 | Mouse | 10.8 |
| NM_008920.4_psr1 | CBP: C-Type Lectin | 12-CTLD + acidic neck | Prg2 [proteoglycan 2 bone marrow] | NM_008920 | Mouse | 12.3 |
| NM_016914.2_psr1 | CBP: C-Type Lectin | 12-CTLD + acidic neck | Prg3 [proteoglycan 3; Eosinophil major basic protein homolog] | NM_016914 | Mouse | 12.1 |
| NM_010048.3_psr1 | CBP: C-Type Lectin | 12-CTLD + acidic neck | DGCR2 DiGeorge syndrome protein C | NM_010048 | Mouse | 11.1 |
| NM_054042.2_psr1 | CBP: C-Type Lectin | 13-IDD | Cd248 [CD248 antigen endosialin] | NM_054042 | Mouse | 11.7 |
| NM_010740.3_psr1_s | CBP: C-Type Lectin | 14-Endosialin | CD93 [C1q receptor; Cd93 antigen] | NM_010740 | Mouse | 10.5 |
| NM_009378.2_psr1 | CBP: C-Type Lectin | 14-Endosialin | Thbd [Thrombomodulin] | NM_009378 | Mouse | 11.6 |
3.7 Statistical Analysis of Gene Expression Data
In order to interpret the DNA microarray data, the raw DNA microarray data files must be imported into a statistical analysis software program. Microsoft® Excel® is the most simple program that could be used, but has limited function. In the model case, GeneSpring® was used; however, there are many commercial programs (e.g. ArrayStar®) and one public domain program (ORIOGEN) available. These programs provide multiple statistical and post-hoc tests, visualization and clustering tools, and often pathway mapping. GeneSpring® can directly import the CEL data generated by the Affymetrix compatible scanner. Many gene expression analysis software tools can also import gene expression data from tab-delimited text files. A portion of a tab-delimited text file from the GLYCOv4 GeneChip® is shown in Table 3. In this case, the data from three biological replicates were combined into a single file.
Table 3.
Example gene expression data outputs for one condition. The gene expression signal, call, and p-values for 10 probe sets are shown for the first 10 genes. Genes that are assigned as present call (P) have p ≤ 0.04. Genes assigned as absent call (A) have p ≥ 0.06. Gene with p-values in-between are assigned marginal (M) calls.
| Probeset ID | Replicate 1 | Replicate 2 | Replicate 3 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Signal | Call | p-value | Signal | Call | p-value | Signal | Call | p-value | |
| NM_013630.2_psr1 | 64 | P | 0.00148 | 73.6 | P | 0.001049 | 100.4 | P | 0.000451 |
| NM_029686.2_psr1 | 37.5 | A | 0.160942 | 16.4 | M | 0.056885 | 25.1 | A | 0.122135 |
| NM_181415.3_psr1 | 7.8 | A | 0.493067 | 1.7 | A | 0.629375 | 14.9 | A | 0.227444 |
| NM_009730.2_psr1 | 566.8 | P | 0.00003 | 428.9 | P | 0.00003 | 404.8 | P | 0.00003 |
| NM_008920.4_psr1 | 32.8 | A | 0.129317 | 30.9 | P | 0.009484 | 31.9 | P | 0.007135 |
| NM_016914.2_psr1 | 6.2 | A | 0.728492 | 3 | A | 0.728492 | 6.9 | A | 0.58937 |
| NM_010048.3_psr1 | 9.5 | A | 0.424197 | 10.3 | A | 0.090421 | 1.6 | A | 0.655316 |
| NM_054042.2_psr1 | 316.8 | P | 0.00003 | 304.5 | P | 0.00003 | 308.4 | P | 0.00003 |
| NM_010740.3_psr1_s | 3.3 | A | 0.990516 | 1.5 | A | 0.870683 | 11 | A | 0.197229 |
| NM_009378.2_psr1 | 5.4 | A | 0.728492 | 2 | A | 0.79298 | 3.7 | A | 0.772556 |
The first step once the data has been imported into the software package is to normalize the raw data between the DNA microarrays. Normalization adjusts for the effects that are due to DNA microarray processing [54]. For example, with two-color DNA microarrays, imbalances in the red and green dyes can occur due to labeling efficiency differences [55]. In GeneSpring®, the researcher needs to select a normalized protocol for the data. GeneSpring® also performs a quality check on the Affymetrix Hybridization Controls, which check for minimum detection levels and titration of concentrations. The default normalization is quartile and it is commonly normalized to the total intensity of the chip, which is often set to the arbitrary value of 500. This method assumes that on average the total RNA level in a cell is constant. Alternatively, one can normalize to the median chip intensity or a particular set of housekeeping genes. Bahr et al. (2009) identified over 100 potential housekeeping genes for CHO cells after reviewing multiple DNA microarray studies [56]. Normalization methods also include linear models, global normalization, LOESS normalization, and subset quantile normalization [57-59]. For Affymetrix CEL files, the steps used are summarization, log transformation, and baseline transformation as described in the GeneSpring® GX Manual.
After data normalization, differentially expressed genes can be identified from the replicates and between the conditions of interest. In the model case, there are three biological replicates for the four conditions, thus data from 12 DNA microarrays was analyzed. Experiment groupings are used to identify sample parameters and the replicates. For example, in the model case ammonium would be a parameter, as well as proline and salt. Table 4 shows the experimental groupings for the model case. Also, a principle component analysis is used by GeneSpring® to check data quality per DNA microarray. DNA microarray data that is of low quality can be discarded if necessary at this step; however, then all the quality control checks are repeated on the new experimental groupings [GeneSpring® GX Manual].
Table 4.
Experimental grouping for the model case to identify replicates for a statistical software package, such as GeneSpring®.
| CEL file names | Parameters | |||
|---|---|---|---|---|
| Replicates | Ammonium | Salt | Proline | |
| Control 1 | Control | 0 | 0 | 0 |
| Control 2 | Control | 0 | 0 | 0 |
| Control 3 | Control | 0 | 0 | 0 |
| Ammonium 1 | Ammonium | 10 | 0 | 0 |
| Ammonium 2 | Ammonium | 10 | 0 | 0 |
| Ammonium 3 | Ammonium | 10 | 0 | 0 |
| Salt 1 | Salt | 0 | 10 | 0 |
| Salt 2 | Salt | 0 | 10 | 0 |
| Salt 3 | Salt | 0 | 10 | 0 |
| Ammonium with Proline 1 | Ammonium Proline | 10 | 0 | 20 |
| Ammonium with Proline 2 | Ammonium Proline | 10 | 0 | 20 |
| Ammonium with Proline 3 | Ammonium Proline | 10 | 0 | 20 |
An ANOVA analysis is used to detect differential expression. The Welch ANOVA test using a Benjamini and Hochberg false discovery rate correction is a fairly robust tool for multiple comparisons (p 0.05) [60]. Many software packages can also conduct numerous types of post-hoc tests on the data (pairwise comparisons). A Tukey Honestly Significant Difference (HSD) post-hoc test is one example that can identify genes that are significantly different between a pair of conditions [GeneSpring® GX Manual]. Table 5 shows an example Tukey post-hoc for an experiment with four conditions. Analyzing these differentially expressed genes for biological relevant comparisons is crucial. For example, in the model case, a direct comparison of the salt-stressed cultures to the ammonium with proline-stressed cultures may not provide any meaningful biological information. However, the other pairwise comparisons are more likely to result in biologically meaningful information. Specifically, comparing the control and ammonium-stressed cultures would provide information on the effects of ammonium on glycosylation genes. Comparing the control and salt-stressed cultures would identify glycosylation genes sensitive to a low salt addition (a mild osmolarity change). Comparing the ammonium-stressed and ammonium-stressed with proline cultures would identify genes sensitive to the proline addition.
Table 5.
Example pairwise comparison of the culture conditions (Tukey post-hoc) – hypothetical data. The ANOVA analysis (p ≤ 0.05) identified 310 of the 1246-glycosylation and housekeeping genes as regulated across the four conditions. The diagonal represents self-comparisons. Values above the diagonal represent the genes that have significantly different expression levels between the paired conditions. Values below the diagonal represent the genes that are not significantly different between the paired conditions.
| Conditions | Control | Condition 1 (10 mM Ammonium) | Condition 2 (10 mM Salt) | Condition 3 (10 mM Ammonium + 20 mM Proline) |
|---|---|---|---|---|
| Control | X | 230 | 70 | 161 |
| Condition 1 (10 mM Ammonium) | 1016 | X | 153 | 125 |
| Condition 2 (10 mM Salt) | 1176 | 1093 | X | 155 |
| Condition 3 (10 mM Ammonium + 20 mM Proline) | 1085 | 1211 | 1091 | X |
Identifying differentially expressed genes and establishing links between these changes and altered biological activity in response to stressful conditions is crucial. Functional annotation can be used to assist with identifying underlying principles. Several databases can be accessed to provide functional annotation such as the Gene Ontology Consortium (GO), Metacyc, and the Kyoto Encyclopedia of Genes and Genomes (KEGG). The Gene Ontology Consortium maintains a database of controlled vocabularies for the description of molecular functions, biological processes and cellular components of gene products, which can be accessed directly from GeneSpring®. Metacyc is a database of non-redundant, experimentally elucidated metabolic pathways. The Metacyc Pathway Tools Omics Viewers allows for gene expression data to be overlaid onto the metabolic pathway. KEGG is collection of manually drawn pathway maps representing the current knowledge of molecular interaction networks, including glycan biosynthesis and metabolism. Additionally, tools like the MAPPFinder software have been used to identify gene ontology with a higher-than-expected proportion in the differentially expressed data [42].
Many different approaches can be taken to identify the biological significance for an observed set of gene expression changes. Cluster analysis, principle component analysis, heat-maps, k-means clustering, and self-organizing maps (SOM) are all common tools [54,61,62].
Hierarchical clustering was used to identify cell-cycle genes that were differentially expressed due to hyperosmolarity [43] and principal components analysis was used to condense the dimensionality for butyrate-treated culture gene expression data [42]. Wit and McClure (2004) reviewed these various tools and provides examples of how each compares [54].
DNA microarray gene expression data is customarily validated using an orthogonal method, with real-time qRT-PCR being the most commonly employed tool. In the recent cell culture DNA microarray literature, the number of gene expression changes that have been validated by real-time qRT-PCR varies from none to six genes [39,42,43,57,63]. Work by Canalas et al. (2006) demonstrated excellent concordance between real-time qRT-PCR and DNA microarray fold change results using the three common real-time qRT-PCR chemistries; calling into question the need for this validation step [64]. However, real-time RT-qPCR does provide lower thresholds of detection and larger assay ranges, which can be used to complement DNA microarray analysis for both highly and rarely expressed mRNA species [64].
Real-time qRT-PCR uses primers to hybridize the mRNA species and amplify a portion of the mRNA, in which either the DNA product or probes fluoresce proportionally to the amount of mRNA in the sample. Gasparic et al. (2010) recently reviewed nine real-time qRT-PCR technologies and provides a description of the mechanism of each [65]. As with DNA microarrays, it is necessary to normalize the gene expression data. For real-time qRT-PCR, housekeeping genes are used to provide this normalization. Often actin, glyceraldehyde 3-phospahte dehydrogenase (GAPDH), and 18S rRNA are used as normalizers for mammalian mRNA [64,65]. Using many DNA microarray data sets, Bahr et al. (2009) identified over 100 housekeeping gene targets for CHO cells due to consistent expression among a variety of culture stresses [56].
To validate DNA microarray gene expression results by real-time qRT-PCR, one must select a real-time qRT-PCR chemistry, design primers to the target mRNA and housekeeping genes, run the reactions, process the data, then compare to the DNA microarray data. Schefe et al. (2006) provide a comprehensive guide to real-time qRT-PCR methods and data analysis [66]. Canalas et al. (2006) provides details for comparing DNA microarray and real-time qRT-PCR results [64].
In the model case, gene expression levels were quantified using SYBR Green I dye chemistry (Molecular Probes, Eugene, OR) [67,68]. Primers for each of the mRNA targets were designed using Primer Express (Applied Biosystems, Foster City, CA). Conventional reverse transcription polymerase chain reactions (RT-PCR) and 2.0% agarose gels were used to confirm primer design, after which visible products on the gels were sequenced to confirm primer specificity. A one step RT-PCR kit (Promega, WI) was used for both conventional RT-PCR and real-time qRT-PCR. The conventional RT-PCR should be performed, as per kit instructions. The real-time qRT-PCR can be performed using a Smart Cycler (Cepheid, Sunnyvale, CA). For the model case, CT values were calculated using the Gene Expression's CT Difference (GED) method [66]. Alternatively, a second derivative analysis CT method [69] outlined by Peters et al. (2003) and Schwarz et al. (2004) [70,71] and normalized as per Pfaffl (2001) [72] can be used. The normalized gene expression levels for the control, ammonium-stressed, salt-stressed, and ammonium-stressed with proline cultures were assessed using Statistical Analysis Software (SAS). An ANOVA analysis (p ≤ 0.05) was used to identify gene expression differences.
3.8 Data Dissemination
Researchers are often required by journals to submit gene expression data to public databases. Gene Expression Omnibus (GEO) at the National Institutes of Health (NIH) in the United States and ArrayExpress at the European Bioinformatics Institute (EBI) part of the European Molecular Biology Laboratory (EMBL) in the United Kingdom, are two well-recognized public gene expression repositories. These public databases offer great opportunities to compare data to other experimental results as well as learn from results of other experiments. Both GEO and ArrayExpress use the Minimum Information About a Microarray Experiment (MIAME) guidelines that specify the minimum information that should be included when describing a microarray experiment. For GEO submission, the types of data input are categorized – including metadata, raw data files, matrix tables, and the platform. The metadata comprises the descriptive information and protocols used. The raw data files are the original, software-generated quantitative results files. For Affymetrix DNA microarray data, the raw data is contained in the CEL files. The matrix table is a spreadsheet that contains the normalized data. The platform is a description of the DNA microarray architecture and probe sequences. Many commercial platforms have already been submitted to GEO; however, if a new platform has been used, the researcher will have to provide this information. As more data becomes available and accessible through these databases, existing models can be updated and improved. The procedure for GEO submission using MIAME can be found online at http://www.ncbi.nlm.nih.gov/geo/info/MIAME.html [73].
4. Concluding Remarks
DNA microarray technologies are extremely useful tools for evaluating how cells respond at the gene expression level to stresses in their culture environment. It is known that the culture environment can greatly impact protein quality, but so far, there has been limited research devoted to evaluating the gene expression levels in response to these culture conditions. While the lack of sequenced genomes hindered previous research, as more genomes continue to be sequenced, the potential for gene expressional analysis using DNA microarrays continues to grow. With DNA microarrays’ ability to look at many different genes at once, researchers can now evaluate a larger scope of cellular responses to specific stresses. This allows for the discovery of more comprehensive patterns, involving not only the genes responsible for the enzymes in the glycosylation pathway but also genes associated with other cellular functions that indirectly influence the protein quality. Additionally, data collections from gene expression studies are now publicly available online, providing researchers with even more data with which to formulate models. With these deeper insights, more effective strategies can be designed to take advantage of various culture parameters known to positively affect glycosylation and other post-translational modifications in an effort to develop methods to improve both the quality and quantity of therapeutic protein yields.
Acknowledgements
This material is based upon work supported by CSREES/USDA, under project number SC-SC-1700287. Any opinions, findings, conclusions or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the view of the USDA. The DNA microarrays and services were provided by Consortium for Functional Glycomics’ Gene Microarray Core Facility funded by NIH NIGMS Grant# GM62116. Additional funded was provided by Clemson University via a URGC Project Completion Grants and the South Carolina COBRE Center of Biomaterials for Tissue Regeneration sponsored by NIH Grant# P20RR021949.
Abbreviations
- CHO cells
Chinese hamster ovary cells
- NS0 cells
a mouse myeloma cell line
- real-time qRT-PCR
real-time quantitative reverse transcription polymerase chain reaction
- PM
perfect match
- MM
mismatch
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hacker DL, De Jesus M, Wurm FM. 25 years of recombinant proteins from reactor-grown cells - where do we go from here? Biotechnol. Adv. 2009;27:1023–1027. doi: 10.1016/j.biotechadv.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 2.Jenkins N, Murphy L, Tyther R. Post-translational modifications of recombinant proteins: Significance for biopharmaceuticals. Mol. Biotechnol. 2008;39:113–118. doi: 10.1007/s12033-008-9049-4. [DOI] [PubMed] [Google Scholar]
- 3.Lim Y, Wong NSC, Lee YY, Ku SCY, Wong DCF, Yap MGS. Engineering mammalian cells in bioprocessing - current achievements and future perspectives. Biotechnol Appl Biochem. 2010;55:175–189. doi: 10.1042/BA20090363. [DOI] [PubMed] [Google Scholar]
- 4.Ozturk SS, Palsson BO. Effects of dissolved oxygen on hybridoma cell growth, metabolism, and antibody production kinetics in continuous culture. Biotechnol. Prog. 1990;6:437–446. doi: 10.1021/bp00006a006. [DOI] [PubMed] [Google Scholar]
- 5.Patel TP, Parekh RG, Poellering BJ, Prior CP. Different culture methods lead to differences in glycosylation of a murine IgG monoclonal antibody. Biochem. J. 1992;285:839–845. doi: 10.1042/bj2850839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maiorella BL, Winkelhake J, Young J, Moyer B, Bauer R, Hora M, Andya J, Thomson J, Patel T, Parekh R. Effect of culture conditions on IgM antibody structure, pharmacokinetics and activity. Biotechnology. 1993;11:387–392. doi: 10.1038/nbt0393-387. [DOI] [PubMed] [Google Scholar]
- 7.Hayter PM, Curling EMA, Gould ML, Baines AJ, Jenkins N, Salmon I, Strange PG, Bull AT. The effect of the dilution rate on CHO cell physiology and recombinant interferon-γ production in glucose-limited chemostat culture. Biotechnol. Bioeng. 1993;42:1007–1085. doi: 10.1002/bit.260420909. [DOI] [PubMed] [Google Scholar]
- 8.Chotigeat W, Watanapokasin Y, Mahler S, Gray PP. Role of enviromental conditions on the expression levels, glycoform pattern and levels of sialyltransferase for hFSH produced by recombinant CHO. Cytotechnology. 1994;15:217–221. doi: 10.1007/BF00762396. [DOI] [PubMed] [Google Scholar]
- 9.Gawlitzek M, Conradt HS, Wagner R. Effect of different cell culture conditions on the polypeptide integrity and N-glycosylation of a recombinant model glycoprotein. Biotechnol. Bioeng. 1995;46:536–544. doi: 10.1002/bit.260460606. [DOI] [PubMed] [Google Scholar]
- 10.Castro PML, Ison AP, Hayter PM, Bull AT. The macrohetrogeneity of recombinant human interferon-γ produced by Chinese hamster ovary cells is affected by the protein and lipid content of the culture medium. Biotechnol Appl Biochem. 1995;21:87–100. [PubMed] [Google Scholar]
- 11.Gu XJ, Xie LZ, Harmon BJ, Wang DIC. Influence of primatone RL supplementation on sialylation of recombinant human interferon-γ produced by Chinese hamster ovary cell culture using serum-free media. Biotechnol. Bioeng. 1997;56:353–360. doi: 10.1002/(SICI)1097-0290(19971120)56:4<353::AID-BIT1>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 12.Chuppa S, Tsai Y-S, Yoon S, Shackleford S, Rozales C, Bhat R, Tsay G, Matanguihan C, Konstantinov K, Naveh D. Fermentor temperature as a tool for control of high-density perfusion cultures of mammalian cells. Biotechnol. Bioeng. 1997;55:328–338. doi: 10.1002/(SICI)1097-0290(19970720)55:2<328::AID-BIT10>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 13.Etchevarry T, Ryll T, Genentech, Inc. Mammalian cell culture process. U.S. Patent 5,705,364. 1998
- 14.Kunkel JP, Jan DCH, Jamieson JC, Butler M. Dissolved oxygen concentration in serum-free continuous culture affects N-linked glycosylation of a monoclonal antibody. J. Biotechnol. 1998;62:55–71. doi: 10.1016/s0168-1656(98)00044-3. [DOI] [PubMed] [Google Scholar]
- 15.Hooker AD, Green NH, Baines AJ, Bull AT, Jenkins N, Strange PG, James DC. Constraints on the transport and glycosylation of recombinant IFN-γ in Chinese hamster ovary and insect cells. Biotechnol. Bioeng. 1999;63:559–572. doi: 10.1002/(sici)1097-0290(19990605)63:5<559::aid-bit6>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 16.Valley U, Nimtz M, Conradt HS, Wagner R. Incorporation of ammonium into intracellular UDP-activated N-acetylhexosamines and into carbohydrate structures in glycoproteins. Biotechnol. Bioeng. 1999;64:401–417. [PubMed] [Google Scholar]
- 17.Kaufmann H, Mazur X, Fussenegger M, Bailey JE. Influence of low temperature on productivity, proteome and protein phosphorylation of CHO cells. Biotechnol. Bioeng. 1999;63:573–582. doi: 10.1002/(sici)1097-0290(19990605)63:5<573::aid-bit7>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 18.deZengotita VM, Abston LR, Schmelzer AE, Shaw S, Miller WM. Selected amino acids protect hybridoma and CHO cells from elevated carbon dioxide and osmolality. Biotechnol. Bioeng. 2002;78:741–752. doi: 10.1002/bit.10255. [DOI] [PubMed] [Google Scholar]
- 19.Chung BS, Jeong YT, Chang KH, Kim JS, Kim JH. Effect of sodium butyrate on glycosylation of recombinant erythropoietin. J. Microbiol. Biotechnol. 2001;11:1087–1092. [Google Scholar]
- 20.Baker KN, Rendall MH, Hills AE, Hoare M, Freedman RB, James DC. Metabolic control of recombinant protein N-glycan processing in NS0 and CHO cells. Biotechnol. Bioeng. 2001;73:188–202. doi: 10.1002/bit.1051. [DOI] [PubMed] [Google Scholar]
- 21.Yang M, Butler M. Effects of ammonia and glucosamine on the heterogeneity of erythropoietin glycoforms. Biotechnol. Prog. 2002;18:129–138. doi: 10.1021/bp0101334. [DOI] [PubMed] [Google Scholar]
- 22.Sung YH, Lee GM. Enhanced human thrombopoietin production by sodium butyrate addition to serum-free suspension culture of bcl-2-overexpressing CHO cells. Biotechnol. Prog. 2005;21:50–57. doi: 10.1021/bp049892n. [DOI] [PubMed] [Google Scholar]
- 23.Jenkins N, Meleady P, Tyther R, Murphy L. Strategies for analysing and improving the expression and quality of recombinant proteins made in mammalian cells. Biotechnol Appl Biochem. 2009;53:73–83. doi: 10.1042/BA20080258. [DOI] [PubMed] [Google Scholar]
- 24.Kim NY, Kim JH, Kim HJ. Effect of low adapted temperature and medium composition on growth and erythropoietin (EPO) production by Chinese hamster ovary cells. Arch. Pharmacal Res. 2005;28:220–226. doi: 10.1007/BF02977719. [DOI] [PubMed] [Google Scholar]
- 25.Yoon SK, Kim SH, Lee GM. Effect of low culture temperature on specific productivity and transcription level of anti-4-1BB antibody in recombinant Chinese hamster ovary cells. Biotechnol. Prog. 2003;19:1383–1386. doi: 10.1021/bp034051m. [DOI] [PubMed] [Google Scholar]
- 26.Clark KJR, Griffiths J, Bailey KM, Harcum SW. Gene-expression profiles for five key glycosylation genes for galactose-fed CHO cells expressing recombinant IL-4/13 cytokine trap. Biotechnol. Bioeng. 2005;90:568–577. doi: 10.1002/bit.20439. [DOI] [PubMed] [Google Scholar]
- 27.Clark KJR, Chaplin FWR, Harcum SW. Temperature effects on product-quality-related enzymes in batch CHO cell cultures producing recombinant tPA. Biotechnol. Prog. 2004;20:1888–1892. doi: 10.1021/bp049951x. [DOI] [PubMed] [Google Scholar]
- 28.Chen PF, Harcum SW. Effects of elevated ammonium on glycosylation gene expression in CHO cells. Metab. Eng. 2006;8:123–132. doi: 10.1016/j.ymben.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 29.Chen PF, Harcum SW. Differential display identifies genes in Chinese hamster ovary cells sensitive to elevated ammonium. Appl. Biochem. Biotechnol. 2007;141:349–359. doi: 10.1007/BF02729072. [DOI] [PubMed] [Google Scholar]
- 30.Wong NSC, Wati L, Nissom PM, Feng HT, Lee MM, Yap MGS. An investigation of intracellular glycosylation activities in CHO cells: Effects of nucleotide sugar precursor feeding. Biotechnol. Bioeng. 2010;107:321–336. doi: 10.1002/bit.22812. [DOI] [PubMed] [Google Scholar]
- 31.Wong DCF, Wong NSC, Goh JSY, May LM, Yap MGS. Profiling of N-glycosylation gene expression in CHO cell fed-batch cultures. Biotechnol. Bioeng. 2010;107:516–528. doi: 10.1002/bit.22828. [DOI] [PubMed] [Google Scholar]
- 32.Jaluria P, Konstantopoulos K, Betenbaugh M, Shiloach J. A perspective on microarrays: Current applications, pitfalls, and potential uses. Microbial Cell Factories. 2007;6 doi: 10.1186/1475-2859-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Redestig H, Costa IG. Detection and interpretation of metabolite-transcript coresponses using combined profiling data. Bioinformatics. 2011;27:I357–I365. doi: 10.1093/bioinformatics/btr231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Androulakis IP, Yang E, Almon RR. Analysis of time-series gene expression data: Methods, challenges, and opportunities. Annu. Rev. Biomed. Eng. 2007;9:205–228. doi: 10.1146/annurev.bioeng.9.060906.151904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu X, Nagarajan H, Lewis NE, Pan S, Cai Z, Liu X, Chen W, Xie M, Wang W, Hammond S, Andersen MR, Neff N, Passarelli B, Koh W, Fan HC, Wang J, Gui Y, Lee KH, Betenbaugh MJ, Quake SR, Famili I, Palsson BO, Wang J. The genomic sequence of the Chinese hamster ovary (CHO)-K1 cell line. Nat. Biotechnol. 2011;29:735–741. doi: 10.1038/nbt.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Korke R, Gatti MD, Lau ALY, Lim JWE, Seow TK, Chung MCM, Hu WS. Large scale gene expression profiling of metabolic shift of mammalian cells in culture. J. Biotechnol. 2004;107:1–17. doi: 10.1016/j.jbiotec.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 37.Wlaschin KF, Seth G, Hu WS. Toward genomic cell culture engineering. Cytotechnology. 2006;50:121–140. doi: 10.1007/s10616-006-9004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nissom PM, Sanny A, Kok YJ, Hiang YT, Chuah SH, Shing TK, Lee YY, Wong KTK, Hu WS, Sim MYG, Philp R. Transcriptome and proteome profiling to understanding the biology of high productivity CHO cells. Mol. Biotechnol. 2006;34:125–140. doi: 10.1385/mb:34:2:125. [DOI] [PubMed] [Google Scholar]
- 39.Gatti MD, Wlaschin KF, Nissom PM, Yap M, Hu WS. Comparative transcriptional analysis of mouse hybridoma and recombinant Chinese hamster ovary cells undergoing butyrate treatment. J. Biosci. Bioeng. 2007;103:82–91. doi: 10.1263/jbb.103.82. [DOI] [PubMed] [Google Scholar]
- 40.Kumar N, Gammell P, Clynes M. Proliferation control strategies to improve productivity and survival during CHO based production culture - A summary of recent methods employed and the effects of proliferation control in product secreting CHO cell lines. Cytotechnology. 2007;53:33–46. doi: 10.1007/s10616-007-9047-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yee JC, Gerdtzen ZP, Hu WS. Comparative transcriptome analysis to unveil genes affecting recombinant protein productivity in mammalian cells. Biotechnol. Bioeng. 2009;102:246–263. doi: 10.1002/bit.22039. [DOI] [PubMed] [Google Scholar]
- 42.Kantardjieff A, Jacob NM, Yee JC, Epstein E, Kok YJ, Philp R, Betenbaugh M, Hu WS. Transcriptome and proteome analysis of Chinese hamster ovary cells under low temperature and butyrate treatment. J. Biotechnol. 2010;145:143–159. doi: 10.1016/j.jbiotec.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 43.Shen DA, Kiehl TR, Khattak SF, Li ZJ, He AQ, Kayne PS, Patel V, Neuhaus IM, Sharfstein ST. Transcriptomic responses to sodium chloride-induced osmotic stress: A study of industrial fed-batch CHO cell cultures. Biotechnol. Prog. 2010;26:1104–1115. doi: 10.1002/btpr.398. [DOI] [PubMed] [Google Scholar]
- 44.Qian YM, Khattak SF, Xing ZZ, He AQ, Kayne PS, Qian NX, Pan SH, Li ZJ. Cell culture and gene transcription effects of copper sulfate on Chinese hamster ovary cells. Biotechnol. Prog. 2011;27:1190–1194. doi: 10.1002/btpr.630. [DOI] [PubMed] [Google Scholar]
- 45.Clarke C, Doolan P, Barron N, Meleady P, O'Sullivan F, Gammell P, Melville M, Leonard M, Clynes M. Predicting cell-specific productivity from CHO gene expression. J. Biotechnol. 2011;151:159–165. doi: 10.1016/j.jbiotec.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 46.Gawlitzek M, Ryll T, Lofgren J, Sliwkowski MB. Ammonium alters N-glycan structures of recombinant tnfr-IgG: Degradative versus biosynthetic mechanisms. Biotechnol. Bioeng. 2000;68:637–646. doi: 10.1002/(sici)1097-0290(20000620)68:6<637::aid-bit6>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 47.Gawlitzek M, Valley U, Wagner R. Ammonium ion and glucosamine dependent increases of oligosaccharide complexity in recombinant glycoproteins secreted from cultivated BHK-21 cells. Biotechnol. Bioeng. 1998;57:518–528. [PubMed] [Google Scholar]
- 48.Zhou WC, Chen CC, Buckland B, Aunins J. Fed-batch culture of recombinant NS0 myeloma cells with high monoclonal antibody production. 1997;55:783–792. doi: 10.1002/(SICI)1097-0290(19970905)55:5<783::AID-BIT8>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 49.Lee YY, Yap MGS, Hu WS, Wong KTK. Low-glutamine fed-batch cultures of 293-hek serum-free suspension cells for adenovirus production. Biotechnol. Prog. 2003;19:501–509. doi: 10.1021/bp025638o. [DOI] [PubMed] [Google Scholar]
- 50.Wong DCF, Wong KTK, Goh LT, Heng CK, Yap MGS. Impact of dynamic online fed-batch strategies on metabolism, productivity and N-glycosylation quality in CHO cell cultures. Biotechnol. Bioeng. 2005;89:164–177. doi: 10.1002/bit.20317. [DOI] [PubMed] [Google Scholar]
- 51.Heller MJ. DNA microarray technology: Devices, systems, and applications. Annu. Rev. Biomed. Eng. 2002;4:129–153. doi: 10.1146/annurev.bioeng.4.020702.153438. [DOI] [PubMed] [Google Scholar]
- 52.Muyal JP, Singh SK, Fehrenbach H. DNA-microarray technology: Comparison of methodological factors of recent technique towards gene expression profiling. Crit. Rev. Biotechnol. 2008;28:239–251. doi: 10.1080/07388550802428400. [DOI] [PubMed] [Google Scholar]
- 53.Larkin JE, Frank BC, Gavras H, Sultana R, Quackenbush J. Independence and reproducibility across microarray platforms. Nat. Methods. 2005;2:337–343. doi: 10.1038/nmeth757. [DOI] [PubMed] [Google Scholar]
- 54.Wit E, McClure J. Statistics for microarrays: Design, analysis, and inference. John Wiley and Sons; West Sussex: 2004. [Google Scholar]
- 55.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 56.Bahr SM, Borgschulte T, Kayser KJ, Lin N. Using microarray technology to select housekeeping genes in Chinese hamster ovary cells. Biotechnol. Bioeng. 2009;104:1041–1046. doi: 10.1002/bit.22452. [DOI] [PubMed] [Google Scholar]
- 57.Khoo SHG, Falciani F, Al-Rubeai M. A genome-wide transcriptional analysis of producer and non-producer NS0 myeloma cell lines. Biotechnol Appl Biochem. 2007;47:85–95. doi: 10.1042/BA20060185. [DOI] [PubMed] [Google Scholar]
- 58.Wu ZJ, Aryee MJ. Subset quantile normalization using negative control features. J. Comput. Biol. 2010;17:1385–1395. doi: 10.1089/cmb.2010.0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30 doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.B B, Y H. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Statist. Soc. B. 1995;57:289–300. [Google Scholar]
- 61.Landgrebe J, Wurst W, Welzl G. Permutation-validated principal components analysis of microarray data. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-4-research0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kantardjieff A, Nissom PM, Chuah SH, Yusufi F, Jacob NM, Mulukutla BC, Yap M, Hu WS. Developing genomic platforms for Chinese hamster ovary cells. Biotechnol. Adv. 2009;27:1028–1035. doi: 10.1016/j.biotechadv.2009.05.023. [DOI] [PubMed] [Google Scholar]
- 63.Wlaschin KF, Hu WS. Engineering cell metabolism for high-density cell culture via manipulation of sugar transport. J. Biotechnol. 2007;131:168–176. doi: 10.1016/j.jbiotec.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 64.Canales RD, Luo YL, Willey JC, Austermiller B, Barbacioru CC, Boysen C, Hunkapiller K, Jensen RV, Knight CR, Lee KY, Ma YQ, Maqsodi B, Papallo A, Peters EH, Poulter K, Ruppel PL, Samaha RR, Shi LM, Yang W, Zhang L, Goodsaid FM. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat. Biotechnol. 2006;24:1115–1122. doi: 10.1038/nbt1236. [DOI] [PubMed] [Google Scholar]
- 65.Gasparic MB, Tengs T, La Paz JL, Holst-Jensen A, Pla M, Esteve T, Zel J, Gruden K. Comparison of nine different real-time PCR chemistries for qualitative and quantitative applications in GMO detection. Anal. Bioanal. Chem. 2010;396:2023–2029. doi: 10.1007/s00216-009-3418-0. [DOI] [PubMed] [Google Scholar]
- 66.Schefe JH, Lehmann KE, Buschmann IR, Unger T, Funke-Kaiser H. Quantitative real-time RT-PCR data analysis: Current concepts and the novel “Gene expression's c(t) difference” Formula. Journal of Molecular Medicine-Jmm. 2006;84:901–910. doi: 10.1007/s00109-006-0097-6. [DOI] [PubMed] [Google Scholar]
- 67.Richards GP, Watson MA, Kingsley DH. A SYBR green, real-time RT-PCR method to detect and quantitate Norwalk virus in stools. Journal of Virological Methods. 2004;116:63–70. doi: 10.1016/j.jviromet.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 68.Hein J, Schellenberg U, Bein G, Hackstein H. Quantification of murine IFN-gamma mRNA and protein expression: Impact of real-time kinetic RT-PCR using SYBR green I dye. Scandinavian Journal of Immunology. 2001;54:285–291. doi: 10.1046/j.1365-3083.2001.00928.x. [DOI] [PubMed] [Google Scholar]
- 69.Luu-The V, Paquet N, Calvo E, Cumps J. Improved real-time RT-PCR method for high-throughput measurements using second derivative calculation and double correction. BioTechniques. 2005;38:287–293. doi: 10.2144/05382RR05. [DOI] [PubMed] [Google Scholar]
- 70.Schwarz G, Baumler S, Block A, Felsenstein FG, Wenzel G. Determination of detection and quantification limits for SNP allele frequency estimation in DNA pools using real time PCR. Nucleic Acids Res. 2004;32 doi: 10.1093/nar/gnh020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peters IR, Helps CR, Batt RM, Day MJ, Hall EJ. Quantitative real-time RT-PCR measurement of mRNA encoding alpha-chain, pIgR and J-chain from canine duodenal mucosa. J. Immunol. Methods. 2003;275:213–222. doi: 10.1016/s0022-1759(03)00056-5. [DOI] [PubMed] [Google Scholar]
- 72.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29 doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Muertter RN, Holko M, Ayanbule O, Yefanov A, Soboleva A. NCBI GEO: Archive for functional genomics data sets - 10 years on. Nucleic Acids Res. 2011;39:D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]