Summary
The redox-regulated chaperone Hsp33 protects bacteria specifically against stress conditions that cause oxidative protein unfolding, such as treatment with bleach or exposure to peroxide at elevated temperatures. To gain insight into the mechanism by which expression of Hsp33 confers resistance to oxidative protein unfolding conditions, we made use of V. cholerae strain O395 lacking the Hsp33 gene hslO. We found that this strain, which is exquisitely bleach-sensitive, displays a temperature-sensitive (ts) phenotype during aerobic growth, implying that V. cholerae suffers from oxidative heat stress when cultivated at 43°C. We utilized this phenotype to select for E. coli genes that rescue the ts phenotype of V. cholerae ΔhslO when overexpressed. We discovered that expression of a single protein, the elongation factor EF-Tu, was sufficient to rescue both the ts and bleach-sensitive phenotypes of V. cholerae ΔhslO. In vivo studies revealed that V. cholerae EF-Tu is highly sensitive to oxidative protein degradation in the absence of Hsp33, indicating that EF-Tu is a vital chaperone substrate of Hsp33 in V. cholerae. These results suggest an “essential client protein” model for Hsp33’s chaperone action in Vibrio in which stabilization of a single oxidative stress-sensitive protein is sufficient to enhance the oxidative stress resistance of the whole organism.
Keywords: Chaperone, Oxidative stress, Redox Regulation, Cysteine, Protein Unfolding
Introduction
The heat shock protein Hsp33 is a highly conserved, redox-regulated chaperone, which has been shown to specifically protect bacteria against a variety of different oxidative stress conditions that are accompanied by protein unfolding. These stress conditions include exposure to hypochlorous acid (HOCl), the active ingredient of household bleach and a known physiological antimicrobial, produced by cells of the innate immune response to kill invading microorganisms (Miller and Britigan, 1997). HOCl is a fast acting oxidant, which directly induces protein unfolding both in vitro and in vivo (Winter et al., 2008). Other physiological oxidants, such as peroxide or nitric oxide, do not cause widespread protein unfolding in organisms and induce activation of Hsp33 only when combined with protein unfolding conditions, such as heat shock treatment (i.e., oxidative heat stress) (Winter et al., 2005). The reason why bacteria require Hsp33 particularly under oxidative protein unfolding conditions is likely due to the fact that enzymes involved in ATP-generation fall victim to oxidative inactivation (Hyslop et al., 1988) causing ATP-dependent chaperones, commonly used to protect against protein aggregation, to lose their in vivo function (Winter et al., 2005). Hsp33, which functions as an ATP-independent chaperone and is activated by oxidative unfolding apparently compensates for this loss of ATP-dependent chaperone activity by protecting hundreds of different proteins against protein aggregation in E. coli (Ilbert et al., 2007, Winter et al., 2008). At this point, it is still unresolved whether the protective role of chaperones such as Hsp33 results from the general decrease in the pool of aggregated proteins, from the protection of a single essential protein whose stress sensitivity dictates the stress sensitivity of the organism, or from something in between.
To investigate how expression of the chaperone Hsp33 confers resistance to oxidative protein unfolding conditions in bacteria, we made use of V. cholerae strain O395 lacking the Hsp33 gene hslO. We found that this strain, which has been previously shown to be highly HOCl-stress sensitive, displays a temperature-sensitive phenotype under aerobic growth conditions, implying that cultivation of V. cholerae at 43°C causes oxidative protein unfolding. We utilized this phenotype to select for E. coli-specific system(s) that compensate for the deletion of Vibrio hslO. We discovered that expression of the E. coli elongation factor EF-Tu fully rescues the temperature-sensitive phenotype of V. cholerae hslO deletions and restores bleach resistance to wild-type levels. In vivo studies revealed that V. cholerae EF-Tu is rapidly degraded in the absence of Hsp33. Expression of E. coli EF-Tu compensates for the lack of Hsp33, suggesting that the cytoprotective effect of the general chaperone Hsp33 in Vibrio comes from guarding a single stress-sensitive protein, EF-Tu, whose presence is essential for the survival of the organism.
Results
V. cholerae Hsp33 null mutants reveal a temperature-sensitive (ts) phenotype
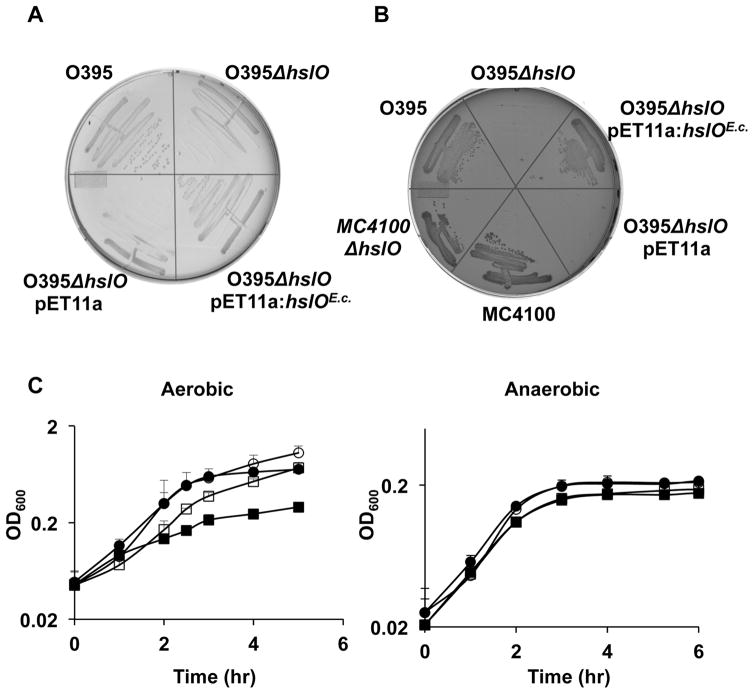
In vitro studies showed that the highly specialized bacterial chaperone Hsp33 contains a dual stress-sensing mechanism, which mediates activation of the chaperone function specifically under oxidative stress conditions that lead to protein unfolding (Ilbert et al., 2007, Winter et al., 2008). In vivo studies confirmed these results and showed that absence of the Hsp33 gene hslO significantly decreases E. coli’s resistance to HOCl stress or oxidative heat stress treatment but does not affect E. coli’s survival at high concentrations of peroxide or at elevated temperatures alone (Winter et al., 2005). It thus came as a surprise when we tested the hslO deletion phenotype in V. cholerae O395 and found this strain to be severely temperature-sensitive (ts) for growth. As shown in Fig. 1A and B, compared to wild-type cells, V. cholerae ΔhslO forms significantly smaller colonies on LB plates and fails to form any colonies on MacConkey plates after 24 h of incubation at 43°C. The temperature sensitivity of hslO null mutants in Vibrio and the finding that Hsp33 functions as an oxidative stress-regulated chaperone in E. coli suggested that heat treatment of V. cholerae either causes or exacerbates oxidative stress conditions that induce the activation of Hsp33, which in turn enhances the survival of V. cholerae at high temperatures. To investigate whether reactive oxygen species (ROS) indeed affect Vibrio’s survival at elevated temperatures, we compared the growth of V. cholerae wild type with that of the hslO deletion mutant in liquid media under both aerobic and anaerobic growth conditions (Fig. 1C). When cultivated under aerobic conditions, we observed a slight growth disadvantage in ΔhslO strains at 37°C and a significant reduction in growth rate at 43°C when compared to the growth of wild type O395. This result was fully consistent with the ts phenotype of this mutant strain on plates. In contrast, however, when we cultivated the same strains under anaerobic conditions, the growth rates of wild type and ΔhslO mutants strains were not significantly different at either 37°C or 43°C (Fig. 1C). These results suggest that at elevated temperatures V. cholerae suffers from oxidative heat stress, which requires activation of Hsp33’s chaperone function for survival. Expression of E. coli Hsp33 in the V. cholerae ΔhslO deletion strain was able to fully complement the ts phenotype of this strain (Fig. 1A and B), excluding significant differences in the activation requirements between the two Hsp33 homologues.
Fig. 1. Aerobically grown V. cholerae ΔhslO strain has ts phenotype.
A and B. Wild-type V. cholerae O395, O395 ΔhslO, or O395 ΔhslO expressing either the empty pET11a plasmid or E. coli Hsp33 from a pET11a plasmid were grown on LB plates (A) or MacConkey plates (B) for 24 h at 43°C. Wild-type E. coli MC4100 and the corresponding MC4100 Delta;hslO mutant strain are shown as controls.
C. V. cholerae O395 (circles) or O395 ΔhslO (squares) were cultivated in LB growth medium at either 37°C (open symbols) or 43°C (filled symbols) in the presence (left panel) or absence (right panel) of air oxygen. Bacterial growth was monitored by optical density measurements at 600 nm.
Identification of E. coli genes that rescue the ts phenotype of O395 ΔhslO
We reasoned that the severe ts phenotype of the O395 ΔhslO mutant strain on MacConkey plates might serve as an in vivo selection system to identify E. coli proteins that, when overexpressed in V. cholerae, protect against oxidative heat stress and, by extension, against HOCl-mediated protein damage. The complementing E. coli gene could encode Hsp33 itself or proteins that function in a way analogous to Hsp33 in parallel pathways. Alternatively, rescuing genes could encode E. coli homologues for important Vibrio Hsp33 substrates, which are either Hsp33-independent in E. coli or become Hsp33-independent in Vibrio simply by increasing their steady state concentrations. Since we were searching for proteins that are potent in rescuing hslO null mutants at relatively low levels of expression, we constructed genomic expression libraries from wild type E. coli MG1655 using either pBR322 (15–20 copies per cell) or pET11a plasmids as expression vectors. We had previously observed that pET11a mediated expression of E. coli Hsp33 in hslO null strains lacking the T7 DNA polymerase provides sufficient Hsp33 levels to allow complementation (Fig. 1A and B). We transformed the genomic library into the O395ΔhslO mutant strain and selected for transformants showing robust growth on MacConkey plates after 24 h incubation at 43°C. All six investigated transformants encoded the E. coli hslO gene. To avoid repeated cloning and identification of the hslO gene, we next constructed a genomic library using chromosomal DNA of the MG1655 ΔhslO deletion mutant WC126, and performed the same selection procedure. We found two independent clones, clone 5 from the pBR322 library and clone 12 from the pET11a library, which conferred a high degree of complementation (Fig. 2A). Cultivation of V. cholerae ΔhslO strain mutants expressing either clone 5 or 12 on MacConkey plates at 43°C yielded colonies that were similar in size to colonies formed by V. cholerae wild-type or by V. cholerae ΔhslO strains expressing E. coli Hsp33 from either pBR322 or pET11a plasmid (Fig. 2A). In contrast, a V. cholerae ΔhslO mutant strain expressing the empty vector failed to form colonies under these conditions. As mentioned previously, Hsp33 protects bacteria against oxidative protein unfolding conditions induced by either high concentrations of peroxide at elevated temperatures (Winter et al., 2005) or by low concentrations of HOCl (Winter et al., 2008). To test whether expression of either one of the two identified E. coli clones rescues the HOCl-sensitive phenotype of the O395 ΔhslO mutant as well, we exposed O395 wild-type, O395 ΔhslO, or the O395 ΔhslO mutant strains expressing either clone 5 or clone 12 to a 10 μM HOCl treatment for 20 min and tested their survival (Fig. 2B). Consistent with earlier studies (Winter et al., 2008), O395 ΔhslO was significantly more sensitive to HOCl treatment than wild type. Importantly, O395 ΔhslO mutant strains expressing either clone 5 or 12 were resistant to HOCl treatment. These results indicate that the gene(s) encoded on these plasmids are capable of protecting O395 ΔhslO against a variety of different stress conditions that cause oxidative protein unfolding.
Fig. 2. E. coli expression library contains clones that rescue ts and HOCl-sensitive phenotypes of O395 ΔhslO mutant.
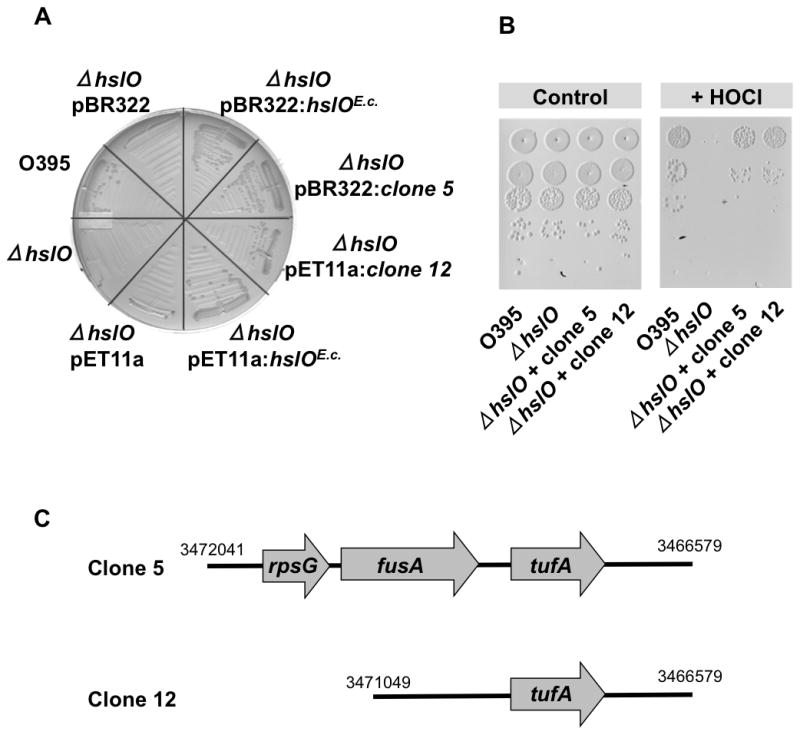
A. E. coli gene expression library from MG1655 ΔhslO mutant strain was constructed in either pET11A or pBR322 plasmids and transformed into the V. cholerae O395 ΔhslO mutant strain. Transformants were cultivated on MacConkey plates for 24 h at 43°C. Two independently identified transformants, clone 5 from the pBR322 library and clone 12 from the pET11a library, were selected for further analysis. Growth of these two strains was tested on MacConkey plates for 24 h at 43°C and compared to O395 wild type, O395 ΔhslO, and O395 ΔhslO expressing the respective empty plasmids.
B. To test the bleach sensitivity of these strains, wild-type O395, O395 ΔhslO, or O395 ΔhslO expressing either clone 5 or clone 12 were cultivated in LB medium until mid-log phase was reached. Cells were washed, resuspended in phosphate buffer, and treated with 10 μM HOCl for 20 min. Cell viability was analyzed by preparing serial dilutions of the cultures and spotting them onto LB plates.
C. Schematic presentation of E. coli genomic sequences with indicated chromosomal positions that were inserted into clone 5 or clone 12.
E. coli EF-Tu expression rescues the ts phenotype of V. cholerae O395 ΔhslO
To investigate which E. coli genes are responsible for rescuing the temperature- and HOCl-sensitive phenotype of the V. cholerae O395 ΔhslO mutant, we sequenced the inserts in clones 5 and 12. While the pBR322:clone 5 contained a sequence spanning three separate genes (rpsG, fusA, and tufA), the pET11a:clone 12 contained only one complete gene (tufA) that encodes elongation factor Tu (EF-Tu) (Fig. 2C). Phenotypical analysis of O395 ΔhslO expressing subclones of pBR322:clone 5 with either rpsG or rpsG/fusA deleted verified that the presence of the E. coli tufA gene, including its upstream regions, is sufficient to rescue the ts phenotype of the O395 ΔhslO mutant strain. Erase-A-Base (Promega) was used to identify the minimal sequence sufficient to complement the ts phenotype of O395 ΔhslO. The shortest sequence capable of rescuing the ts phenotype contained 405 bases upstream of the tufA gene as well as the complete tufA gene. This result is in excellent agreement with previous studies showing that the promoter region of tufA is about 400 bases upstream of the start codon (Zengel and Lindahl, 1990). We concluded from these studies that expression of a single protein, E. coli EF-Tu, is necessary and sufficient to rescue the ts phenotype and, by extension, the HOCl-sensitive phenotype of a V. cholerae mutant strain lacking Hsp33. We noted that E. coli EF-Tu was not massively overexpressed; rather, its expression levels were comparable to the endogenous level of EF-Tu in wild-type V. cholerae (see below).
Hsp33 is essential for maintaining high levels of soluble EF-Tu in V. cholerae
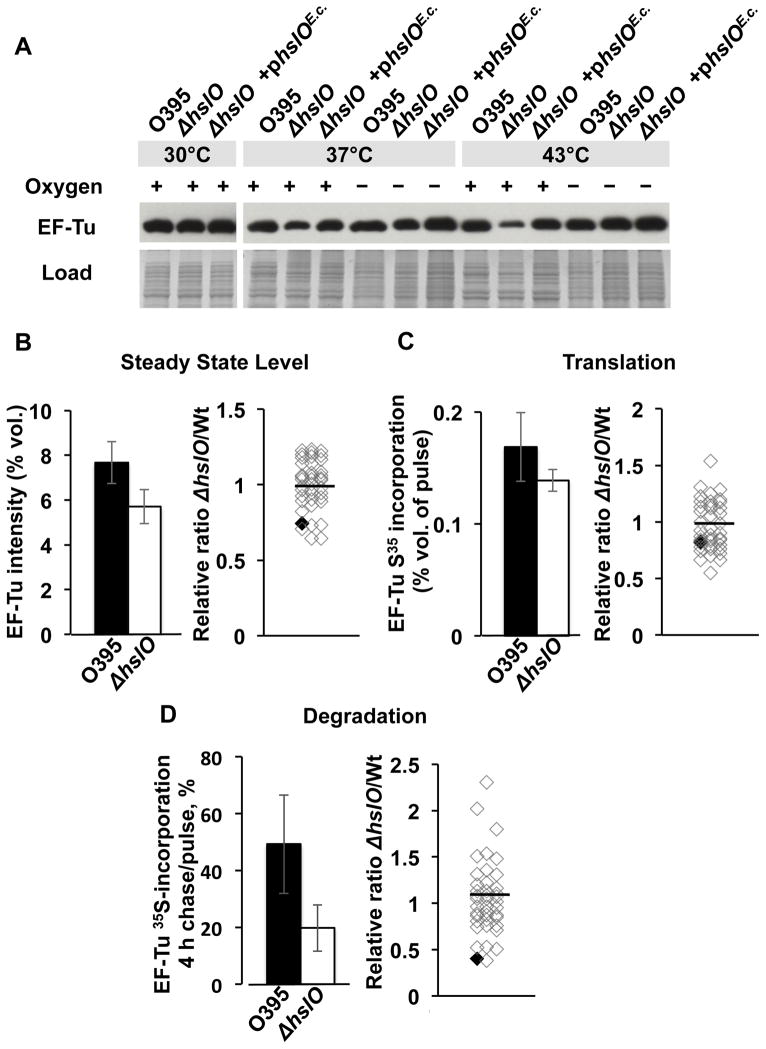
The elongation factor EF-Tu is one of the most abundant proteins in the bacterial cytosol and is essential for cell growth (Pedersen et al., 1978). To begin to understand how overexpression of E. coli EF-Tu can rescue the temperature-sensitive growth defect of the O395 ΔhslO mutant strain on plates and in cultures, we compared the levels of endogenous EF-Tu in V. cholerae and O395 ΔhslO mutant strains under both non-stress and heat shock conditions in the absence of additional E. coli EF-Tu. We found that lack of Hsp33 did not cause a noticeable change in the steady state concentration of endogenous Vibrio EF-Tu at 30°C, but led to a reproducible decrease in EF-Tu levels at 37°C and a very substantial decrease at 43°C when compared to EF-Tu levels in either wild type or the O395ΔhslO mutant expressing E. coli Hsp33 from a plasmid (Fig. 3A). When we cultivated the strains anaerobically, however, no difference in the steady state levels of EF-Tu was detected at any temperature (Fig. 3A). This result is entirely consistent with the lack in phenotype of hslO null mutants in Vibrio under anaerobic growth conditions (Fig. 1C) and serves to show that elevated temperatures alone do not affect EF-Tu levels in V. cholerae. These results suggest that under aerobic heat shock conditions, presence of the redox-regulated chaperone Hsp33 is necessary to maintain EF-Tu at cellular levels that are sufficient for cell growth. Note that we saw no differences in the steady state levels of EF-Tu in E. coli wild-type vs. E. coli ΔhslO strains at 43°C, which is consistent with the fact that the E. coli ΔhslO deletion strains are not temperature-sensitive for growth (Winter et al., 2005).
Fig. 3. Hsp33 protects V. cholerae EF-Tu against protein degradation.
A. Wild-type O395, O395 ΔhslO mutant, or O395 ΔhslO mutant encoding E. coli hslO on a pBR322 plasmid (ΔhslO + phslOE.c.) were cultivated in LB medium at the indicated temperature in the presence and absence of air oxygen. Cells were harvested at mid-log growth and lysed. Steady state levels of EF-Tu were visualized using Western blot analysis with polyclonal antibodies against E. coli EF-Tu.
B–D. V. cholerae wild-type and V. cholerae ΔhslO mutant strains were cultivated in minimum MOPS medium supplemented with all amino acids except methionine and cysteine. Cells were pulsed for 2 min with radioactive 35S-methionine, flushed with cold unlabeled methionine, and chased for 4 h. The cell lysates were prepared and proteins were separated by 2D PAGE and scanned for 35S incorporation. The error bars represent standard errors from 4 individual experiments.
B. Left Panel: To compare the steady state EF-Tu levels in V. cholerae wild type (black bar) and ΔhslO mutant (white bar), the relative spot intensity of EF-Tu on Coommassie-stained 2D gels was determined for both strain backgrounds. Right Panel: The relative spot intensity of the 40 most abundant protein spots (see Fig. S2A) was determined in both ΔhslO and wild type V. cholerae and compared. Individual proteins are represented by open diamonds. EF-Tu is indicated as black diamond.
C. Left Panel: To compare the 35S incorporation into EF-Tu from V. cholerae wild type and ΔhslO mutant, the relative spot intensity of EF-Tu on the respective autoradiographs was determined. Right Panel: The relative ratio of 35S-incorporation into the 40 most abundant protein spots in ΔhslO and wild type V. cholerae was determined. Individual proteins are represented by open diamonds. EF-Tu is indicated with a black diamond.
D. Left Panel: To determine the rates of protein degradation, 35S incorporation after 4 h of chase relative to 35S incorporation after 2 min pulse was calculated for EF-Tu in V. cholerae wild type and ΔhslO mutant. Right Panel: Protein degradation rates were determined for the 40 most abundant protein spots in both ΔhslO and wild type V. cholerae and compared. Individual proteins are represented by open diamonds. EF-Tu is indicated with a black diamond.
Absence of Hsp33 leads to accelerated degradation of EF-Tu
Changes in the steady state levels of a protein are either the result of decreased rates of transcription and translation and/or are caused by increased rates of proteolysis. To assess the mRNA levels of tufA in V. cholerae and V. cholerae ΔhslO strains, we performed RT-PCR under aerobic conditions at three different temperatures: 30°C, 37°C, and 43°C. No significant difference in tufA transcript levels was observed (Fig. S1), arguing against the possibility that Hsp33 either directly or indirectly affects the expression of tufA. Our results strongly suggested that Hsp33 acts at the post-transcriptional level, presumably by protecting EF-Tu against premature degradation.
To determine the proteolytic stability of endogenous EF-Tu in the presence and absence of Hsp33, we conducted pulse-chase experiments combined with 2D gel electrophoresis. We performed these experiments at non-stress temperatures to exclude that major differences in the growth rates of V. cholerae wild-type and ΔhslO deletion strains affect our data analysis. We reproducibly identified the same 300 protein spots on 2D gels and autoradiographs and used them as reference spots for our analysis (see Experimental procedures for details). Analysis of the Coommassie stained 2D gels confirmed our previous observations and showed a 26% reduction in EF-Tu steady state levels in the O395 ΔhslO mutant strain as compared to wild-type O395 at non-stress temperatures (Fig. 3B, left panel). Westernblot analysis failed to detect any fragments of EF-Tu. When we compared the ratio of EF-Tu steady state levels in O395 wild type and the corresponding ΔhslO deletion mutant to the 40 most abundant protein spots (Fig. S2A), we found the ratio of EF-Tu to be significantly below the mean (Figs 3B, right panel and S3), suggesting that Hsp33 affects EF-Tu-levels rather specifically.
Analysis of the autoradiographs revealed that EF-Tu translation was not significantly different in wild-type and mutant cells, confirming that Hsp33 has no effect on transcription or translation of EF-Tu (Fig. 3C, Fig. S2B). In contrast, we found that the rate of EF-Tu degradation in wild-type and ΔhslO mutant strains was dramatically different. We detected almost 50% of the original 35S-label after 4 h of pulse in EF-Tu isolated from wild-type cells, whereas only 20% of the original label was detected in EF-Tu isolated from cells lacking Hsp33. These results indicate that EF-Tu was degraded 2.5-fold faster in the hslO deletion strain than in wild-type cells (Fig. 3D; Fig. S2C, D). Compared to the 40 most abundant protein spots on the 2D gel, EF-Tu showed again the largest difference in degradation rates between wild type and the hslO deletion mutant (Fig. 3D, right panel, and Fig. S3). These results strongly suggest that EF-Tu is a key Hsp33 client protein, a conclusion, which is consistent with global protein-protein interaction studies in E. coli that showed that Hsp33 is an interaction partner of EF-Tu (Butland et al., 2005).
V. cholerae EF-Tu is exquisitely sensitive to oxidative stress treatment
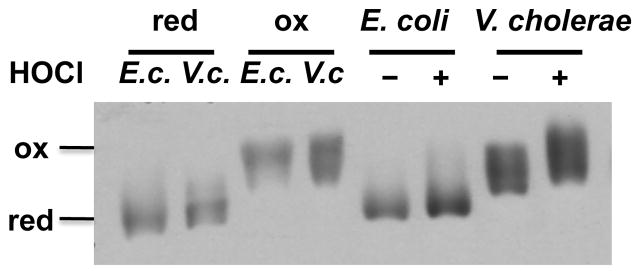
Our studies demonstrated that during aerobic growth of V. cholerae, presence of Hsp33 is required to maintain the stability of the EF-Tu protein. The fact that plasmid-driven expression of E. coli EF-Tu is sufficient to complement the ts phenotype of the O395 ΔhslO mutant furthermore suggested that V. cholerae EF-Tu might exhibit higher oxidative stress sensitivity than E. coli EF-Tu. It has been previously shown that E. coli EF-Tu, although apparently insensitive to peroxide-mediated thiol modifications, quickly responds to HOCl stress treatment with the reversible modification of at least one of its three cysteine residues (Leichert et al., 2008). To compare the in vivo redox status of E. coli and V. cholerae EF-Tu before and after HOCl treatment, we performed differential thiol-trapping experiments using MC4100 and O395 strains. Both strains were cultivated in LB medium at 37°C to mid-log phase. Cell aliquots were removed before and 20 min after HOCl treatment, and lysed in the presence of 10% TCA to prevent any further thiol oxidation (Zander et al., 1998). All reduced cysteine thiols were irreversibly alkylated with N-ethylmaleimide (NEM), whereas all reversibly oxidized cysteines were reduced with DTT and subsequently labeled with the 500 Da thiol-specific alkylation reagent 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS). The latter labeling step introduces a 500 Da molecular mass to every cysteine residue that was originally oxidized in vivo. This mass addition significantly slows the migration of AMS-labeled proteins and allows direct visualization of the in vivo redox status of proteins on one-dimensional SDS-PAGE. We observed a striking difference in the in vivo redox status of V. cholerae and E. coli EF-Tu, particularly in exponentially growing bacteria. While E. coli EF-Tu was almost completely reduced during logarithmic growth, the majority of V. cholerae EF-Tu was already partially oxidized (Fig. 4, compare lanes 5 and 7). Treatment of V. cholerae with sublethal concentrations of HOCl shifted almost all of the endogenous EF-Tu into the fully oxidized species. Comparative analysis of NEM-trapped samples on reducing and non-reducing gels suggested the formation of both inter- and intramolecular disulfide bonds in HOCl-treated V. cholerae EF-Tu (Fig. S4). In contrast, the same treatment of E. coli cells led only to a partial oxidation of E. coli EF-Tu and no visible intermolecular disulfide bond formation (Fig. 4, Fig. S4). These results suggest that V. cholerae EF-Tu is highly susceptible to oxidation and appears to be exposed to substantial levels of reactive oxygen species during aerobic exponential growth even at non-stress temperatures.
Fig. 4. V. cholerae EF-Tu is exquisitely sensitive to oxidative thiol modifications.
To determine the redox status of EF-Tu in vivo, E. coli MC4100 and V. cholerae O395 were grown in LB medium at 37°C under aerobic conditions until mid-log phase was reached. Cells were either left untreated or were treated with 3 mM HOCl for 20 min. Samples were taken and cysteines were labeled with NEM, followed by the reduction of all oxidized thiols and the labeling of all newly reduced cysteines with the 500 Da thiol-alkylating molecule AMS. Addition of AMS molecules is visualized as migration difference on SDS-PAGE, whose extent directly reflects the number of in vivo oxidized cysteines. To define the migration behavior of fully NEM-labeled EF-Tu (equivalent to reduced species) and fully AMS labeled EF-Tu (equivalent to completely oxidized species), aliquots of non-stressed MC4100 or O395 cells (indicated by E.c. and V.c., respectively) were reduced with DTT and labeled exclusively with either NEM (lanes 1 and 2) or AMS (lanes 3 and 4). Proteins were separated on SDS-PAGE and EF-Tu was visualized with Western blot analysis using antibodies against E. coli EF-Tu.
Expression of E. coli EF-Tu affects V. cholerae EF-Tu levels in vivo
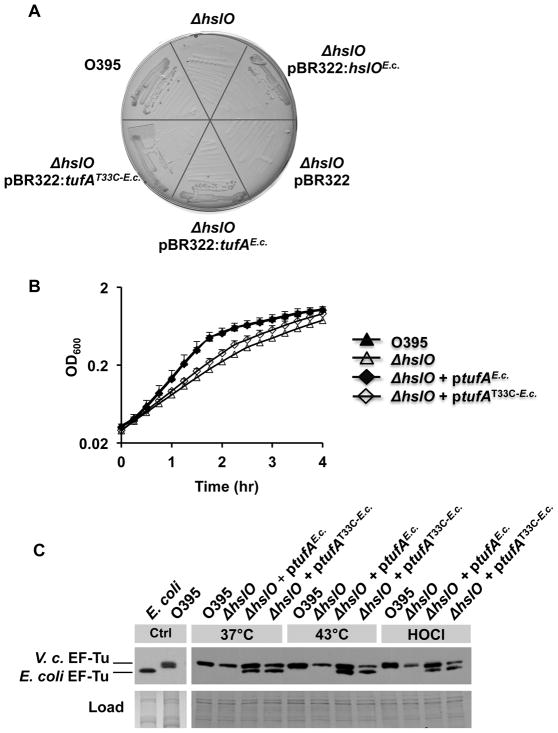
Our results suggested that V. cholerae EF-Tu is a highly oxidative stress-sensitive protein whose loss in steady state levels in the absence of Hsp33 is compensated by expressing the potentially more oxidative stress-resistant E. coli EF-Tu homologue. Amino acid sequence comparison between E. coli and V. cholerae EF-Turevealed that in addition to the three cysteine residues that are present in the two EF-Tu homologues, V. cholerae EF-Tuencodes one additional cysteine residue that is located at position 33 (Cys33) (Fig. S5). To elucidate whether this cysteine is responsible for the increased oxidative stress sensitivity of V. cholerae EF-Tu, we generated a variant of E. coli EF-Tu that carries an additional cysteine residue at this position. For these experiments we used E. coli EF-Tu since we were unable to express detectable levels of V. cholerae EF-Tufrom plasmid constructs in either V. cholerae or E. coli despite the use of various expression vectors. We expressed the E. coli EF-TuT33C variant in the Vibrio O395 ΔhslO mutant strain to test its ability to rescue its ts phenotype. We expected that any increase in oxidative stress sensitivity or overall decrease in EF-Tu stability caused by introducing the T33C mutation into E. coli EF-Tushould lead to lower steady state levels of EF-Tu and thus to a lower capacity of this mutant protein to rescue the ts or bleach-sensitive phenotype of the V. cholerae O395 ΔhslO mutant. As shown in Fig. 5A, the O395 ΔhslO mutant expressing E. coli EF-TuT33C formed slightly smaller colonies on MacConkey plates at 43°C than O395 wild-type or O395 ΔhslO mutant strains expressing wild-type E. coli EF-Tufrom the same plasmid. Moreover, monitoring the aerobic growth of these strains in LB medium at 43°C revealed that expression of the E. coli EF-TuT33C variant was only partially able to rescue the growth defect of O395 ΔhslO (Fig. 5B). We then decided to analyze the levels of soluble E. coli EF-Tuand E. coli EF-TuT33C variant upon plasmid-mediated expression in O395 ΔhslO at 43°C, which was made possible by the fact that V. cholerae EF-Tu and E. coli EF-Tu differ significantly in their migration behavior on SDS-PAGE and hence can be distinguished even when co-expressed (Fig. 5C). We did not find any significant difference in the cellular levels of the two E. coli EF-Tu variants, suggesting that the simple presence of additional EF-Tu might not be sufficient to rescue the ts phenotype of O395 ΔhslO (Fig. 5C). What we did notice, however, was a significant difference in the levels of endogenous Vibrio EF-Tu. Whereas expression of wild type E. coli EF-Tuin O395 ΔhslO mutant strains raised the endogenous EF-Tu levels to those observed in wild-type O395 at both 37°C and 43°C, expression of the E. coli EF-TuT33C variant did not affect the endogenous levels of V. cholerae EF-Tu (Fig. 5C). These results suggested that wild-type E. coli EF-Tu but not E. coli EF-TuT33C confers stability to Vibrio EF-Tu, thus increasing its steady state levels and potentially contributing to the enhanced stress survival observed in these bacteria. We obtained very similar results when we analyzed the steady state levels of V. cholerae EF-Tu in response to bleach treatment at 37°C. While expression of either Hsp33 or E. coli EF-Tu significantly stabilized the levels of endogenous EF-Tu in the presence of HOCl, absence of Hsp33 or presence of the E. coli EF-TuT33C mutant led to substantially increased bleach-mediated degradation of endogenous EF-Tu. These results thus suggest that expression of E. coli EF-Tu functionally replaces Hsp33 at least in part by protecting V. cholerae EF-Tu against oxidative protein degradation. Furthermore, our study demonstrates that the oxidative stress sensitivity of the single bacterial protein EF-Tu is sufficient to determine the cellular survival of V. cholerae at elevated temperatures and in the presence of the physiological antimicrobial bleach.
Fig. 5. E. coli EF-Tu compensates for lack of Hsp33 by protecting V. cholerae EF-Tu against oxidative protein degradation.
A. V. cholerae O395 wild type, O395 ΔhslO mutant, or O395 ΔhslO expressing the empty pBR322 vector, E. coli Hsp33, E. coli EF-Tu,or the E. coli EF-TuT33C variant were cultivated on MacConkey plates for 24 h at 43°C.
B. V. cholerae O395 wild type (black triangles), O395 ΔhslO mutant (white triangles), and O395 ΔhslO expressing either E. coli EF-Tu(black diamonds) or the E. coli EF-TuT33C variant (white diamonds)were cultivated in LB medium at 43°C aerobically. Bacterial growth was monitored by optical density measurements at 600 nm.
C. To analyze the steady state levels of EF-Tu in these bacterial strains, the different strains were cultivated in LB medium at either 37°C or 43°C until mid-log phase was reached. To evaluate the effects of HOCl on cellular EF-Tu levels, the 37°C cultures were split and either left untreated or incubated with 3 mM HOCl for 20 min. Cell aliquots were taken and the proteins were separated by SDS-PAGE. EF-Tu was visualized by Western blot using antibodies against E. coli EF-Tu.
Discussion
Hsp33 is a highly specialized chaperone, which appears to be selectively activated by protein unfolding conditions in the presence of elevated ROS levels, such as experienced by organisms during hypochlorous acid stress or oxidative heat stress (Winter et al., 2008). Absence of Hsp33 in bacteria exposed to these specific stress conditions causes a substantial growth disadvantage, allowing us to use the hslO deletion phenotype as an indicator for oxidative protein unfolding in vivo. Here we report the surprising finding that deletion of the Hsp33 gene hslO in V. cholerae, a mutant strain with a high sensitivity to HOCl stress (Winter et al., 2008), displays a temperature-sensitive (ts) growth defect. Importantly, we found that the ts phenotype was fully abrogated when the mutant bacteria were cultivated under anaerobic conditions. One reasonable explanation is that V. cholerae is exposed to significant ROS production during aerobic growth and requires Hsp33 as an alternative chaperone once it encounters protein unfolding induced by stress conditions, such as elevated temperatures.
The severe ts phenotype of V. cholerae hslO deletion mutants provided us with the opportunity to use a genetic approach to shed light on the in vivo mechanism of Hsp33’s chaperone action. We reasoned that by searching for genes in addition to hslO that are capable of rescuing the ts phenotype of V. cholerae ΔhslO mutants, we might discover alternative E. coli chaperones or antioxidant systems that are able to replace Hsp33 under oxidizing protein unfolding stress conditions. Alternatively, we might identify bacterial proteins whose high sensitivity to oxidative protein unfolding causes the observed phenotype and can be compensated by an increase in their steady state levels. We independently selected two EF-Tu clones that rescued both the temperature-sensitive and bleach-sensitive phenotypes of V. cholerae lacking Hsp33. EF-Tu promotes binding of aminoacyl-tRNA to the ribosome and therefore allows peptide chain elongation during protein biosynthesis (Thompson et al., 1986). Previous in vivo studies provided evidence that E. coli EF-Tu is a redox-sensitive protein that shows elevated levels of thiol oxidation during aerobic growth and undergoes additional oxidative thiol modifications in response to HOCl treatment (Leichert et al., 2008). Moreover, exposure of E. coli cells to near-lethal HOCl stress conditions or oxidative heat shock caused EF-Tu’s aggregation, suggesting that excessive thiol modifications might induce protein unfolding (Winter et al., 2008). Finally, it was found that decreasing the levels of EF-Tu by deleting the tufA gene increased E. coli’s bleach sensitivity (Leichert et al., 2008). Our studies in V. cholerae were fully consistent with the results in E. coli. Yet, our finding that EF-Tu’s cysteines become oxidized simply by growing Vibrio cholerae under aerobic conditions suggested an even higher oxidation sensitivity of Vibrio EF-Tu as compared to E. coli EF-Tu. Absence of the redox-regulated chaperone Hsp33 then leads to the premature degradation of EF-Tu, which causes a decrease in growth rates and, by a yet to be defined mechanism, a significant increase in bleach-sensitivity.
At this point, it is unclear how expression of E. coli EF-Tu confers enhanced bleach resistance to V. cholerae. The simplest and most straightforward explanation of our results would be that E. coli wild-type EF-Tu has increased oxidative stress resistance, hence functionally replacing V. cholerae EF-Tu in protein translation and promoting E. coli’s recovery after bleach stress by rapidly resuming protein translation. Introduction of the additional Cys33 would increase EF-Tu’s oxidative stress sensitivity and hence abrogate this protective function. However, our studies led to a very unexpected finding: we showed that presence of E. coli EF-Tu not only increased the steady-state levels of V. cholerae EF-Tu during aerobic growth but significantly stabilized Vibrio EF-Tu both during heat stress and upon short-term treatment with bleach. Hence the beneficial effects of E. coli EF-Tu expression were less likely simply due to an increase in the translation rate of V. cholerae EF-Tu but appeared to involve stabilization of Vibrio EF-Tu towards oxidative protein degradation. This conclusion was also consistent with our analysis of protein translation in V. cholerae strains lacking Hsp33, which appeared to not be affected by the decreased EF-Tu levels. It has long been known that even during exponential growth EF-Tu molecules outnumber ribosomes by a factor of seven (Furano, 1975, Pedersen et al., 1978). This finding has fueled the idea that EF-Tu plays more than one role in the cell. For instance, it has been demonstrated that EF-Tu polymerizes with other proteins to form filamentous, actin-like structure that function to maintain cell shape in E. coli and Bacillus subtilis (Beck, 1979, Defeu Soufo et al., 2010). Moreover, previous in vitro studies showed that purified elongation factor EF-Tu protects thermally unfolding citrate synthase, a commonly used in vitro chaperone substrate, against protein aggregation and supports the refolding of citrate synthase upon return to non-stress temperatures (Caldas et al., 1998, Kudlicki et al., 1997). This potential chaperone function of E. coli EF-Tu might compensate for the lack of Hsp33 and hence stabilizes V. cholerae EF-Tu. We were unable to find increased stabilization or decreased aggregation in response to heat or bleach stress for any other Vibrio protein(s) in hslO deletion mutants expressing E. coli EF-Tu, suggesting that EF-Tu might act specifically with the Vibrio EF-Tu pool, for instance by forming intermolecular dimers (Weijland and Parmeggiani, 1994). Another proposed function of EF-Tu includes an antioxidant scavenger role to buffer oxidants through the use of its ten methionine residues (Luo and Levine, 2009). This scavenging function of EF-Tu has been proposed to decrease the levels of ROS in vivo and hence protect oxidation-sensitive proteins against oxidative damage. As the E. coli EF-TuT33C variant has the same number of methionines and is expressed to the same extent, this non-specific antioxidant function appears to play only a minor role in conferring bleach resistance in V. cholerae. Future studies are clearly needed to address these fundamental questions regarding EF-Tu’s alternative in vivo functions.
In summary, our study revealed that the stress sensitivity of a whole organism is determined in large part by the stress sensitivity of a single essential protein. In the case of Vibrio, this single essential protein appears to be EF-Tu. The stabilization of ET-Tu by the chaperone Hsp33 is sufficient to significantly enhance the stress resistance of the whole organism. It remains now to be determined what essential protein(s) are the “weakest links” in other organisms under the same or other stress conditions. Given the many known stress conditions, which vary in effects and biological targets, it is likely to be different proteins for different stress conditions, justifying the wide substrate specificity of molecular chaperones.
Experimental procedures
Strain and growth condition
The V. cholerae and E. coli strains used in this study can be found in Table 1. Strains were cultivated in Luria-Bertani (LB) medium at the indicated temperatures. Ampicillin (100 μg/ml) was added to those cultures that contained pET11a or pBR322 plasmids. Growth on MacConkey agar at 43°C was used to select for temperature-sensitive phenotypes.
Table 1.
Bacterial strains and plasmids used in this study
| Strain | Relevant genotype | Plasmid | Reference |
|---|---|---|---|
| JW370 | V. cholerae O395 WT | Winter et al. (2008) | |
| JW371 | O395 ΔhslO | Winter et al. (2008 | |
| WC022 | JW371 | pET11a | This study |
| WC025 | JW371 | pET11a:hslOE.c. | This study |
| WC038 | JW371 | pET11a:clone 12 | This study |
| WC028 | JW371 | pBR322 | This study |
| WC159 | JW371 | pBR322:hslOE.c. | This study |
| WC037 | JW371 | pBR322:clone 5 | This study |
| WC157 | JW371 | pBR322:tufAE.c. | This study |
| WC187 | JW371 | pBR322:tufAT33C-E.c. | This study |
| WC126 | MG1655 | Lab collection | |
| WC127 | WC126 hslO::kan | Lab collection | |
| BB7222 | MC4100 | Winter et al. (2008) | |
| JW176 | BB7222 hslO::kan | Winter et al. (2008) |
Preparation of genomic overexpression library
Genomic DNA from the E. coli wild-type strain MG1655 or the ΔhslO deletion strain JW176 was prepared using the GenElute Bacterial Genomic DNA kit (Sigma-Aldrich). The genomic DNA (2 μg) was partially digested with 0.05–0.2 units of BfuCI (New England Biolabs) in a total volume of 50 μl for 10 min at 37°C. Digested products were analyzed on 1% agaroase gels and showed fragment sizes ranging between 1 and 8 kb. After heat inactivation of enzyme, the DNA fragments were ligated into linearized pBR322/BamH1 or pET11a/BamH1 plasmid vectors using T4 ligase (New England Biolabs). The overexpressing plasmids were transformed into ultracompetent XL10-Gold cells (Stratagene) following the manufacturer’s protocol. Colony forming units of transformants were counted the next day to calculate the size of the libraries (1×105 colonies for pBR322:ΔhslO library; 5×104 colonies for pET11a:ΔhslO library). The transformants of each library were combined and the plasmids were purified using Wizard Plus SV Minipreps kit (Promega). The plasmids were then transformed into V. cholerae ΔhslO mutant strain. The transformants were plated on MacConkey agar and incubated for 24 h at 43°C to select for clones that rescue the ts phenotype of the V. cholerae ΔhslO mutant strain. Transformants that formed healthy looking colonies on plates were re-streaked on MacConkey plates and grown at 43°C. Plasmids of 12 transformants per library were purified and re-transformed into V. cholerae ΔhslO mutant to eliminate the possibility of mutations in the strain background.
HOCl survival assay
To determine the HOCl stress resistance, bacterial stains were cultivated in LB media at 37°C until OD600 of 0.4–0.5 was reached. Due to the reactivity of HOCl, cells were harvested, washed twice with 83 mM sodium phosphate buffer, pH 7.0, and resuspended in the same buffer. The cell density in each sample was normalized to 2×108 cells per ml and treated with the indicated concentrations of sodium hypochlorite (Sigma-Aldrich). After 20 min incubation at room temperature, the treated cells were diluted 1:10 into 5-fold concentrated LB medium to quench the remaining HOCl (Winter et al., 2008). Serial 10-fold dilutions of treated cells were prepared and spotted onto LB plates. The colony-forming units after overnight incubation at 37°C were counted and used for determination of cell survival.
Pulse-chase labeling and 2D gel electrophoresis
V. cholerae O395 wild type and V. cholerae ΔhslO were cultivated in MOPS minimal medium supplemented with 0.2% glucose and all amino acids except methionine and cysteine for 24 h at 37°C. Cells were then diluted 1:80 into fresh media and grown at 35°C until OD600 = 0.4 – 0.5 was reached. Then, 15 μCi/ml radioactive 35S-methionine (Easytag Expre35S Protein Labeling Mix, PerkinElmer) was added to each culture for 2 min (i.e., pulse) followed by the addition of 2.7 mM unlabeled methionine (i.e., chase). Aliquots of 1.8 ml cells were collected immediately after the pulse as well as 1, 2, and 4 h during the chase. All samples were washed twice with ice-cold 60 mM KCl buffer and lysed in DAB buffer (6 M Urea, 200 mM Tris-HCl pH8.5, 10 mM EDTA, and 0.5% w/v SDS). To determine the protein amount in the cell lysates, the DC Protein Assay Kit (Bio-Rad) was applied using BSA as standard. 90 μg of protein from each sample were then pelleted using trichloroacetic acid precipitation and re-dissolved in 450 μl of loading buffer (7 M urea, 2 M thiourea, 1% [w/v] Serdolit MB-1, 1% [w/v] dithiothreitol, 4% [w/v] Chaps, and 0.5% [v/v] Pharmalyte 3–10). The 2D gel electrophoresis, staining of the gels and autoradiography were performed as previously described (Leichert and Jakob, 2004).
Image and data analysis
The protein pattern on the stained 2D gels and autoradiographs were compared and analyzed using the Delta2D 3.6 Software (Decodon). Spot detection, background correction and normalization were performed according to the software’s instructions. Spot matching and alignments across the autoradiographs and stained gels of at least 4 independent pulse-chase experiments were performed. A master fusion gel, which retained all spots located on all individual images (Fig. S2) was generated for spot labeling, visualization and cross-reference between gels. With the built-in settings of Delta2D, the spot quality, pixel intensity (i.e., volume) and % volume (volume of individual spot over total volume of all spots) for each protein spot was determined and exported as a spreadsheet table. To compare the steady state concentration and translation of EF-Tu within a 2 minute time frame in V. cholerae and V. cholerae ΔhslO, we calculated the mean % volume of EF-Tu as well as of the 40 most abundant protein spots on the Coomassie-stained 2D gels or autoradiographs (Fig. S2). To analyze the protein degradation rates in V. cholerae and V. cholerae ΔhslO, we divided the % volume of each spot on the autoradiograph by the % volume of the respective spot on the Coommassie stained 2D gel and determined the fold decrease of 35S incorporation from the 2 min pulse to 4 h of chase. The mean degradation rate for EF-Tu and the each of the 40 pre-selected spots was calculated from four independent pulse-chase experiments (Fig. S3). Standard errors were calculated and are shown in the figures.
Differential in vivo thiol trapping with NEM and AMS
Bacterial strains were cultivated in LB media at 37°C until OD600 of 0.4–0.5 was reached. Then, 3 mM HOCl was added to the medium directly and incubation was continued for 20 min. Before and after the stress treatment, aliquots of 1 ml were taken and acidified with trichloroacetic acid (TCA) to a final concentration of 10%. After 30 min of incubation on ice, precipitated proteins were pelleted by centrifugation (13, 000 rpm, 20 min, 4°C). The protein pellet was resuspendend in DAB buffer (6 M Urea, 200 mM Tris-HCl pH8.5, 10 mM EDTA, and 0.5% w/v SDS) supplemented with 100 mM N-ethylmaleimide (NEM) to irreversibly alkylate all reduced cysteines. Samples were incubated for 30 min at 25°C. The proteins were again precipitated with TCA to remove any unbound NEM, and pelleted by centrifugation. For differential thiol trapping with AMS, protein pellets were resuspended in DAB buffer supplemented with 10 mM DTT to reduce all in vivo oxidized cysteines, and incubated for 1 h at 25°C. Excess DTT was removed by TCA precipitation and centrifugation. All newly accessible cysteines were then modified with 10 mM of the thiol-specific alkylation reagent 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS), which adds 500 Da mass to every modified cysteine. As control, we also prepared fully NEM-labeled proteins, which represent the fully reduced species and fully AMS-labeled proteins, which represent the fully oxidized protein species. Cell aliquots were taken, precipitated with TCA and resuspended in DAB buffer supplemented with 10 mM DTT to reduce all in vivo thiol modifications. After incubation of the samples for 1 h at 25°C, proteins were precipitated with TCA, centrifuged and resuspended in DAB buffer supplemented with either 100 mM NEM or 10 mM AMS. Proteins were separated on SDS-PAGE and EF-Tu was visualized by western blot analysis using polyclonal antibodies against EF-Tu (provided by Dr. Beckwith).
Supplementary Material
Acknowledgments
We thank Drs. J. Bardwell, R. Bender, V. DiRita, and M. Chapman for many helpful discussions. We thank Dr. J. Beckwith for providing us with polyclonal antibodies against E. coli EF-Tu. We thank Drs. Dana Reichmann and Angela Walker for conducting the identification of our proteins by mass spectrometry. W.-Y. W. was supported by the NIH Cellular and Molecular Biology Training Grant T32-GM007315. This project was supported by the National Institutes of Health grant GM065318 to U.J.
References
- Beck BD. Polymerization of the bacterial elongation factor for protein synthesis, EF-Tu. Eur J Biochem. 1979;97:495–502. doi: 10.1111/j.1432-1033.1979.tb13137.x. [DOI] [PubMed] [Google Scholar]
- Butland G, Peregrin-Alvarez JM, Li J, Yang W, Yang X, Canadien V, et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- Caldas TD, El Yaagoubi A, Richarme G. Chaperone properties of bacterial elongation factor EF-Tu. J Biol Chem. 1998;273:11478–11482. doi: 10.1074/jbc.273.19.11478. [DOI] [PubMed] [Google Scholar]
- Defeu Soufo HJ, Reimold C, Linne U, Knust T, Gescher J, Graumann PL. Bacterial translation elongation factor EF-Tu interacts and colocalizes with actin-like MreB protein. Proc Natl Acad Sci U S A. 2010;107:3163–3168. doi: 10.1073/pnas.0911979107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furano AV. Content of elongation factor Tu in Escherichia coli. Proc Natl Acad Sci U S A. 1975;72:4780–4784. doi: 10.1073/pnas.72.12.4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyslop PA, Hinshaw DB, Halsey WA, Jr, Schraufstatter IU, Sauerheber RD, Spragg RG, et al. Mechanisms of oxidant-mediated cell injury. The glycolytic and mitochondrial pathways of ADP phosphorylation are major intracellular targets inactivated by hydrogen peroxide. J Biol Chem. 1988;263:1665–1675. [PubMed] [Google Scholar]
- Ilbert M, Horst J, Ahrens S, Winter J, Graf PC, Lilie H, Jakob U. The redox-switch domain of Hsp33 functions as dual stress sensor. Nat Struct Mol Biol. 2007;14:556–563. doi: 10.1038/nsmb1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudlicki W, Coffman A, Kramer G, Hardesty B. Renaturation of rhodanese by translational elongation factor (EF) Tu. Protein refolding by EF-Tu flexing. J Biol Chem. 1997;272:32206–32210. doi: 10.1074/jbc.272.51.32206. [DOI] [PubMed] [Google Scholar]
- Leichert LI, Gehrke F, Gudiseva HV, Blackwell T, Ilbert M, Walker AK, et al. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc Natl Acad Sci U S A. 2008;105:8197–8202. doi: 10.1073/pnas.0707723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichert LI, Jakob U. Protein thiol modifications visualized in vivo. PLoS Biol. 2004;2:e333. doi: 10.1371/journal.pbio.0020333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Levine RL. Methionine in proteins defends against oxidative stress. FASEB J. 2009;23:464–472. doi: 10.1096/fj.08-118414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RA, Britigan BE. Role of oxidants in microbial pathophysiology. Clin Microbiol Rev. 1997;10:1–18. doi: 10.1128/cmr.10.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen S, Bloch PL, Reeh S, Neidhardt FC. Patterns of protein synthesis in E. coli: a catalog of the amount of 140 individual proteins at different growth rates. Cell. 1978;14:179–190. doi: 10.1016/0092-8674(78)90312-4. [DOI] [PubMed] [Google Scholar]
- Thompson RC, Dix DB, Karim AM. The reaction of ribosomes with elongation factor Tu.GTP complexes. Aminoacyl-tRNA-independent reactions in the elongation cycle determine the accuracy of protein synthesis. J Biol Chem. 1986;261:4868–4874. [PubMed] [Google Scholar]
- Weijland A, Parmeggiani A. Why do two EF-Tu molecules act in the elongation cycle of protein biosynthesis? Trends Biochem Sci. 1994;19:188–193. doi: 10.1016/0968-0004(94)90018-3. [DOI] [PubMed] [Google Scholar]
- Winter J, Ilbert M, Graf PC, Ozcelik D, Jakob U. Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell. 2008;135:691–701. doi: 10.1016/j.cell.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J, Linke K, Jatzek A, Jakob U. Severe oxidative stress causes inactivation of DnaK and activation of the redox-regulated chaperone Hsp33. Mol Cell. 2005;17:381–392. doi: 10.1016/j.molcel.2004.12.027. [DOI] [PubMed] [Google Scholar]
- Zander T, Phadke ND, Bardwell JC. Disulfide bond catalysts in Escherichia coli. Methods Enzymol. 1998;290:59–74. doi: 10.1016/s0076-6879(98)90007-6. [DOI] [PubMed] [Google Scholar]
- Zengel JM, Lindahl L. Mapping of two promoters for elongation factor Tu within the structural gene for elongation factor G. Biochim Biophys Acta. 1990;1050:317–322. doi: 10.1016/0167-4781(90)90188-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.