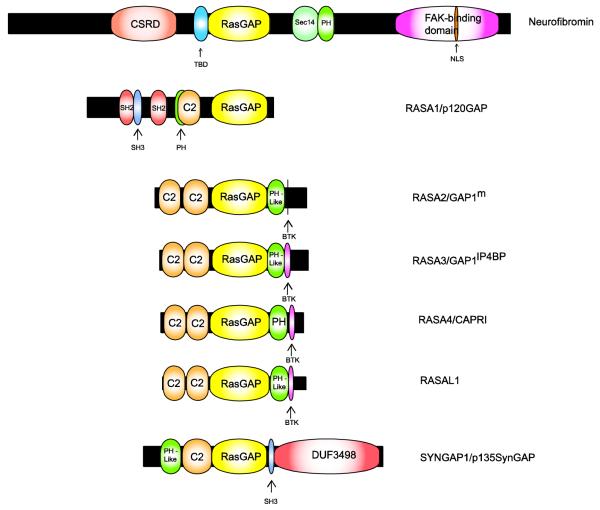
Fig. 2.
Schematic illustrating the functional domains present in neurofibromin and comparing the structure of neurofibromin to that of other Ras GAPs. The lengths of each protein (indicated by the black bars) and domains within each Ras GAP (indicated by the colored expansions) are scaled to the actual number of amino acids in each. See the text for a detailed explanation of known functions of the domains contained within neurofibromin. Domain designations are as follows: CSRD, cysteine/serine-rich domain; TBD, tubulin-binding domain; RasGAP, Ras GTPase-activating protein; Sec14, Sec14-homology domain; PH, pleckstrin homology domain that can target protein to appropriate cellular location and bind inositol phosphates and other proteins; NLS, nuclear localization signal; SH2, domain that binds phosphotyrosine-containing ligands via two surface pockets (a phosphotyrosine pocket and a hydrophobic binding pocket); SH3, domain that binds proline-rich ligands with moderate affinity and selectivity (particularly PxxP motifs); C2, calcium-dependent membrane-targeting module that binds a variety of substrates including phospholipids, inositol polyphosphates and intracellular proteins; BTK, Bruton tyrosine kinase-like motif capable of binding zinc (always follows a PH domain); DUF3498, domain of unknown function.