Figure 1.
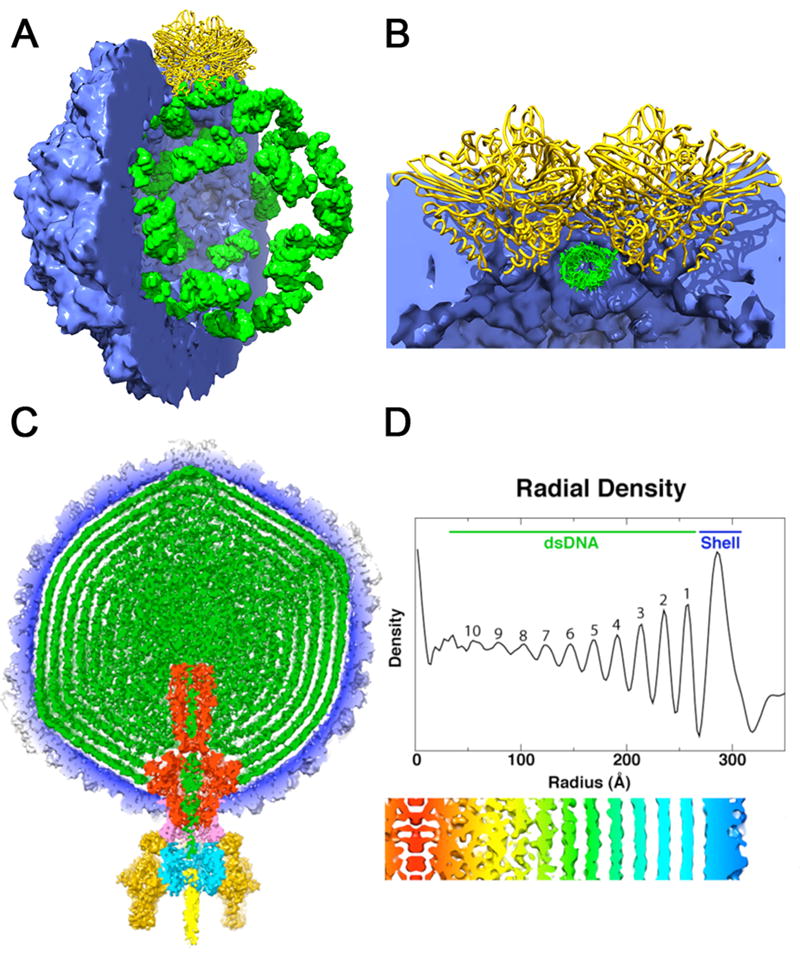
Structures adopted by encapsidated nucleic acid. (a) Packaging of cellular RNA in the metal free FHV virus like particle. The crystal structure coordinates were used to compute density maps for the protein shell (blue) and bound RNA (green). Half of the capsid has been cut away to reveal the dodecahedral arrangement of the ordered duplex RNA segments bound by a coat protein dimer at the icosahedral 2-fold axes (yellow). (b) Close up view of the subunit-RNA interaction looking down the duplex RNA axis and perpendicular to the particle 2-fold. The RNA structure is imposed by the coat protein at the 2-folds to create a molecular switch in the formation of the quasi-equivalent T=3 particle. (c) A 60Å thick central cross-section of the P22 virion showing the concentric layered organization of the packaged dsDNA genome (green). The map is segmented to highlight individual components: coat protein gp5 (blue), gp1 (red), gp4 (pink), gp9 (dark yellow), gp10 (cyan), gp26 (yellow), and pilot proteins (green) [36*]. Here the capsid is a ready made container for the genome and the dsDNA doesn’t play an essential role in particle formation. (d) Radial density plot of the spherically averaged phage reconstruction shows at least ten distinct layers of dsDNA, separated by about 22 Å, exist within the capsid shell. A thin, central section from a map 12-fold averaged about the portal axis and aligned with the radial density plot above it, shows an enlarged view of the DNA layers (red-to-blue, low-to-high radii).
