Abstract
Archaeosine (G+) is found at position 15 of many archaeal tRNAs. In Euryarchaeota, the G+ precursor, 7-cyano-7-deazaguanine (preQ0), is inserted into tRNA by tRNA-guanine transglycosylase (arcTGT) before conversion into G+ by ARChaeosine Synthase (ArcS). However, many Crenarchaeota known to harbor G+ lack ArcS homologs. Using comparative genomics approaches, two families that could functionally replace ArcS in these organisms were identified: 1) GAT-QueC, a two-domain family with an N-terminal glutamine amidotransferase class-II domain fused to a domain homologous to QueC, the enzyme that produces preQ0; 2) QueF-like, a family homologous to the bacterial enzyme catalyzing the reduction of preQ0 to 7-aminomethyl-7-deazaguanine. Here we show that these two protein families are able to catalyze the formation of G+ in a heterologous system. Structure and sequence comparisons of crenarchaeal and euryarchaeal arcTGTs suggest the crenarchaeal enzymes have broader substrate specificity. These results led to a new model for the synthesis and salvage of G+ in Crenarchaeota.
The 7-deazaguanosine nucleosides queuosine (Q) and archaeosine (G+) are two of the most highly modified nucleosides found in tRNA (1). While sharing a common core structure and a significant portion of their biosynthetic pathway (2) (Figure 1A), Q and G+ are segregated phylogenetically and are located in different regions of the tRNA; Q is found in the tRNA of Bacteria and Eukarya at position-34 (the wobble position) in tRNAs decoding NAC/U codons, while G+ is found only in Archaea at position-15 in the dihydrouridine loop (D-loop). Consistent with its position in the anticodon, Q has a role in modulating codon–anticodon binding efficiency (3), while the presence of the positively charged formamidine group of G+ is thought to be important in structural stabilization of the tRNA through electrostatic interactions with the anionic phosphates (4, 5). Computational studies show that G+ can also participate in structural stabilization via strengthening of the hydrogen bonding between the G15-C48 Levitt base pair (4, 5), however neither mechanism has been tested experimentally.
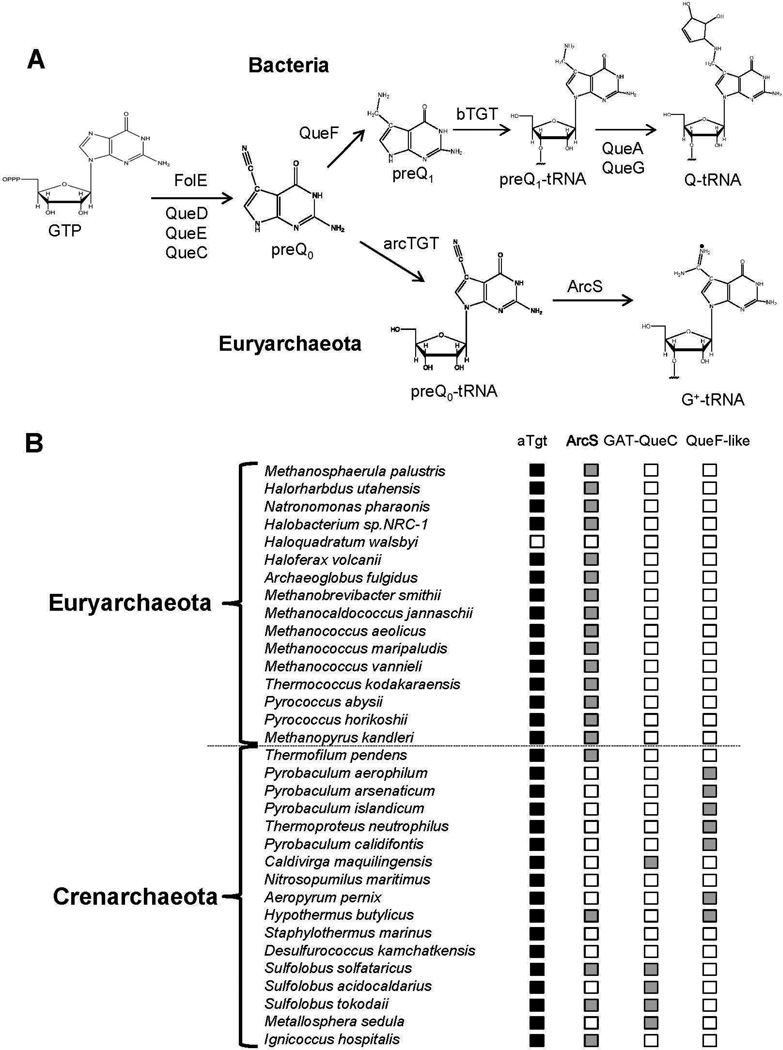
Figure 1. G+ biosynthesis.
A) Experimentally validated G+ and Q biosynthesis steps in Bacteria and Euryarchaeota. B) Phylogenetic distribution of aTGT, ArcS, GAT-QueC and QueF-like in the two archaeal phyla. Filled boxes denote the presence of the gene in the corresponding organism; empty box denotes its absence.
GTP cyclohydrolase I, the first biosynthetic enzyme in the folate/biopterin pathways, is also the first enzyme in the Q/G+ pathways (6), which is followed by the QueD, QueE and QueC enzymes in both Bacteria (7, 8 ) and Archaea (9) to produce the advanced intermediate 7-cyano-7-deazaguanine (preQ0) (Figure 1A). In Archaea, preQ0 is inserted directly into tRNA by a tRNA-guanine transglycosylase (arcTGT, EC 2.4.2.29) (10, 11), encoded by the tgtA gene (10, 11) (Figure 1A). In bacteria, preQ0 is first reduced to 7-aminomethyl-7-deazaguanine (preQ1) by QueF (EC 1.7.1.13) (12) before insertion in substrate tRNAs by a bacterial type TGT (bTGT, EC 2.4.2.29) encoded by the tgt gene (13) (Figure 1A). PreQ1 is further modified on the tRNA to Q in two subsequent enzymatic steps (14, 15).
A recently discovered ATP-independent amidinotransferase, ARChaeosine Synthase or ArcS, catalyzes the final step in the G+ pathway, the conversion of preQ0-tRNA to G+-tRNA, in Euryarchaeota (16) (Figure 1A). No ArcS homolog could be identified in most sequenced Crenarchaeota with the exception of a few Sulfolobii sp. Thermophilus pendens, Hypothermus butylicus, Ignisphaera aggregans and Ignecocci (16) (Figure 1B). However crenarchaeal tRNAs contain G+ (4, 17, 18). The amidino group of G+ must therefore be introduced by other non-homologous enzyme families in these organisms, and here we identify the candidate “missing enzymes” with a combination of comparative genomics and experimental approaches.
RESULTS AND DISCUSSION
G+ is one of the rare archaeal specific tRNA modifications found quasi-universally along the archaeal tree. ArcTGT is found in all Archaea sequenced to date with the exception of the extreme halophile Haloqadratum walsbyi (Figure 1B). Analysis of bulk tRNA extracted from H. walsbyi showed that G+ was indeed absent in this organism (Supplemental Figure 1A), reinforcing arcTGT as a signature enzyme for the G+ pathway. ArcS, however, is not universally distributed: while all sequenced Crenarchaeota contain tgtA genes, the majority lack arcS homologs (Figure 1B). Specific organisms lacking arcS such as Sulfolobus acidocaldarius or Pyrobaculum islandicum are known to contain G+ (4, 17), and we confirmed this for another Pyrobaculum species, Pyrobaculum calidifontis JCM 11548 (Supplemental Figure 1B). This suggests that amidotransferase enzymes responsible for amidation of the nitrile group of the preQ0 precursor are yet to be identified in Crenarchaeota.
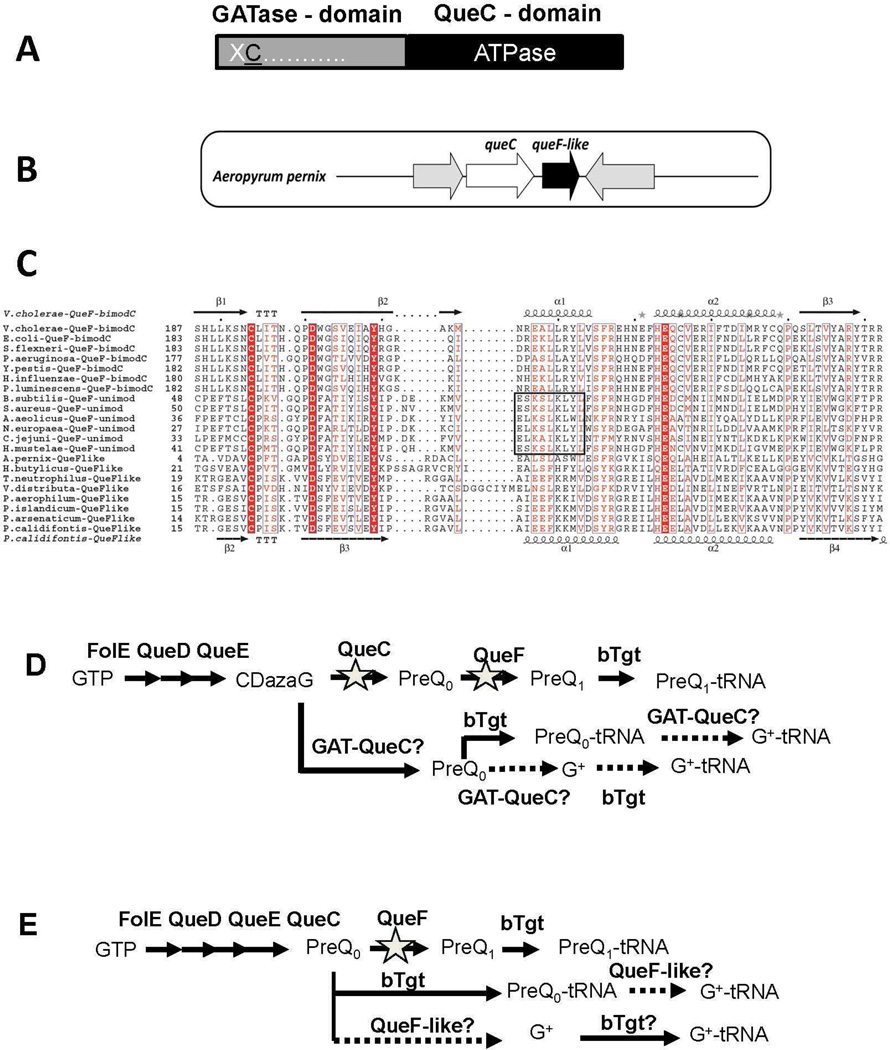
We observed that QueC proteins from several Crenarchaeota are much larger than those from most other Archaea (470 residues instead of 270) because of the presence of an additional N-terminal domain homologous to proteins of the glutamine amidotransferase class-II (GATase) family (Figure 2A and Supplemental data 1). This domain generally catalyzes an ammonia group transfer from glutamine to the appropriate substrate (19). This fused protein family, named here GAT-QueC, is therefore a natural candidate for the missing crenarchaeal enzyme family that would transfer an amido group to the nitrile of preQ0. However, GAT-QueC homologs are not found in all Crenarchaeota that lack ArcS (Figure 1B). We identified another gene family, queF-like (Supplemental data 1), with a member that physically clusters in Aeropyrum pernix with the queC gene (Figure 2B) and encodes a protein family homologous to QueF, the NADPH-dependent enzyme that catalyzes the reduction of preQ0 to preQ1 in bacteria (12) (Figure 1A). QueF are Tunneling-fold enzymes (20) characterized by the QueF motif (E(S/L)K(S/A)hK(L/Y)(Y/F/W) where h is a hydrophobic amino acid), which provides key residues that are proposed to bind the cofactor NADPH. QueF enzymes fall into two subfamilies (12): unimodular QueF enzymes comprised of a single T-fold domain harboring both the QueF motif and the substrate binding pocket, and bimodular QueF enzymes comprised of two weakly homologous tandem T-fold domains with the QueF motif and the substrate binding residues lying separately in the N- and C-terminal domains, respectively (12). To compare the QueF-like family with both QueF subfamilies, we generated homology models for all QueF-like sequences (from eight crenarchaea) and several unimodular and bimodular QueF sequences, and superposed these models with the crystal structures of bimodular V. cholerae QueF (PDB ID 3BP1, (21)) and of unimodular B. subtilis QueF (Swairjo and Iwata-Reuyl, unpublished data) . The predicted QueF-like structures were most similar to the C-terminal T-fold domain of bimodular QueFs (e.g., versus V. cholerae QueF C-terminal half, the r.m.s.d. is 1.5–3.1 Å over 89–107 Cα atoms, 16–22% identity). A structure-based multisequence alignment of QueF-like with unimodular QueF and the C-terminal half of bimodular QueF revealed that residues of the QueF preQ0 binding pocket including Cys55, Tyr70 and Glu97 (in B. subtilis QueF residue numbers) as well as Asp62, which interacts with the nitrogen atom of the substrate cyano group (Swairjo and Iwata-Reuyl unpublished data), are strictly conserved in QueF-like proteins (Figure 2C). The alignment also shows that QueF-like proteins lack the putative signature NADPH-binding motif of QueF (Figure 2C). This led us to propose that the QueF-like proteins found in Archaea are also enzymes that recognize and act on preQ0, but instead of using NADPH to reduce the cyano group, they perform an amidation of preQ0 to form G+.
Figure 2. Analysis of the GAT-QueC and QueF-like protein families.
A) Two domain organization of GAT-QueC enzymes; B) Physical clustering of queF-like with queC in A. pernix; C) Structure-based multisequence alignment of QueF and QueF-like proteins. Invariant residues of the substrate binding pocket are highlighted in red. The QueF motif in unimodular QueF is boxed. Secondary structure elements from the V. cholerae QueF crystal structure and from the P. calidifontis QueF-like homology model are shown above and below the sequences, respectively; D) Design of E. coli test strain; in the E. coli K12 MG1655 strain, the queF and queC were deleted, and the resulted deletion strain was transformed with an expression plasmid containing GAT-queC from S. solfataricus (SSO0016) cloned behind a PBAD promoter. E) Design of the E. coli test strain; queF was deleted in E. coli K12 MG1655, and the resulting deletion strain was transformed with an expression plasmid containing queF-like from P. calidifontis (Pcal_0221) cloned behind a PBAD promoter.
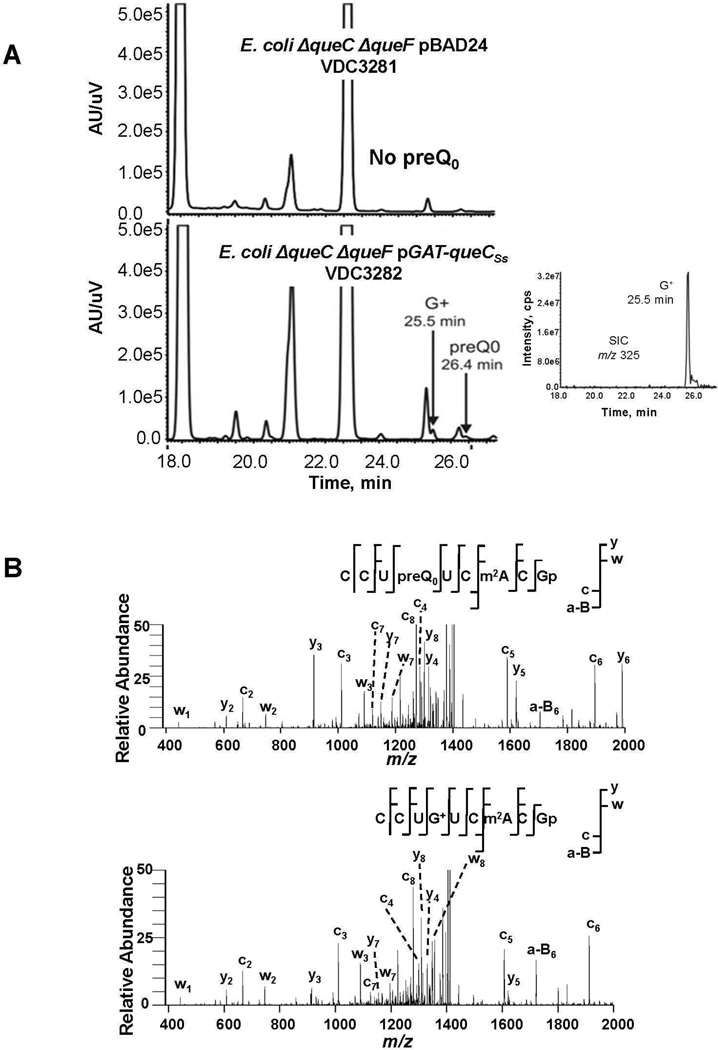
Most crenarchaeal genomes encode a fused GAT-QueC protein or a QueF-like protein (with a clear inverse distribution of the two protein families) (Figure 1B). No good genetic model organisms were available to directly test the function of these two protein families in Archaea. Indeed the only Crenarchaeota with available genetics tools, S. solfataricus (see (9) for review), encodes both a regular ArcS and a GAT-QueC homolog and no genetic tools are available for any organism encoding a QueF-like homolog. We therefore developed heterologous systems based on the common features between Q and G+ synthesis (Figure 1A) to test whether archaeal GAT-QueC and QueF-like proteins could synthesize G+ in E. coli. In one case, a ΔqueC ΔqueF E.coli strain was transformed with a pBAD24 derivative expressing the GAT-QueC gene from S. solfataricus, SSO0016 (Figure 2D). In the other, a ΔqueF E.coli strain was transformed with a pBAD24 derivative expressing the queF-like gene from P. calidifontis, Pcal_0221 (Figure 2E). tRNA was extracted from the different strains grown in Luria Broth (LB) with 0.2% arabinose, digested and dephosphorylated to generate the ribonucleosides for LC/MS/MS analysis. As shown in Figure 3A, tRNA extracted from the ΔqueC ΔqueF strain transformed with an empty vector control (VDC3281) contains no Q or preQ0, as expected, because the precursor pathway has been disrupted. Peaks at 25.5 min and 26.4 min corresponding to G+ (MH+ 325 m/z) and preQ0 nucleoside (MH+ 308 m/z), respectively, were detected in tRNA extracted from the strain expressing SSO0016 (VDC3282). Similarly, the ΔqueF strain transformed with empty pBAD24 (VDC3367) accumulated preQ0 in tRNA as expected (12) (Supplemental Figure 2). The same strain transformed with a derivative expressing Pcal_0221 (VDC3368) contained both preQ0 and G+ (Supplemental Figure 2).
Figure 3. tRNA analysis extracted from the E. coli derivative strains.
A) Analysis of modified nucleosides extracted from E. coli ΔqueF ΔqueC derivative strains. The UV traces at 254 nm and the extraction ion chromatograms (inset) for 325 m/z are shown. The UV chromatogram of the bulk tRNA extracted from VDC3282 showed the G+ peak eluted at 25.5 min and preQ0 peak eluted at 26.4 min. The UV chromatogram of the bulk tRNA extracted from VDC3281 (the negative control) showed no preQ0 peak. B) LC-MS/MS of the RNase T1 digestion products from tRNAAsp purified from the ΔqueF strain. (top) CID of the m/z 1453 digestion product eluting at 30.6 min (Supplemental Figure 4). The detected a-B, c-, w- and y-type ions consistent with the sequence of CCUpreQ0UCm2AGp are identified in this mass spectrum. (bottom) CID of the m/z 1462 digestion product eluting at 29.4 min (Supplemental Figure 4). The detected a-B, c, w- and y-type ions consistent with the sequence of CCUG+UCm2AGp are identified in this mass spectrum.
To identify the positions of preQ0 and G+ in these two strains (VDC3367 and VDC3368), tRNAAsp was purified and sequenced by RNase T1 digestion LC-MS/MS analysis (22, 23). The tRNAAsp purified from strain VDC3367 was found to have a single digestion product ([M-H]− 2907), detected at the 2- (m/z 1453.25) and 3- (m/z 968.75) charge states, that was not expected from the published wild type sequence of this tRNA (24) and was consistent with having the sequence CCUpreQ0UCm2ACGp (Supplemental Figure 3). tRNAAsp purified from strain VDC3368 had the same unique digestion product as seen in strain VDC3367 and an additional product ([M-H]− 2925), detected at the 2- (m/z 1461.92) and 3- (m/z 974.42) charge states (Supplemental Figure 4). This new digestion product is consistent with the sequence CCUG+UCm2ACGp, and was also detected as the cyclic phosphate (Supplemental Figure 4).
Collision-induced dissociation (CID) tandem mass spectrometry was used to confirm these sequence assignments. The assigned CID spectra of m/z 1453 and m/z 1462 were consistent with the sequences CCUpreQ0UCm2AGp and CCUG+UCm2AGp, respectively (Figure 3B). CID spectra from all of the RNase T1 digestion products also were analyzed and were consistent with tRNAAsp wild-type sequence except for preQ0 at position 34 from both the VDC3367 and VDC3368 strains and G+ at position 34 found only in the VDC3368 strain tRNA (Supplemental Figure 5) .
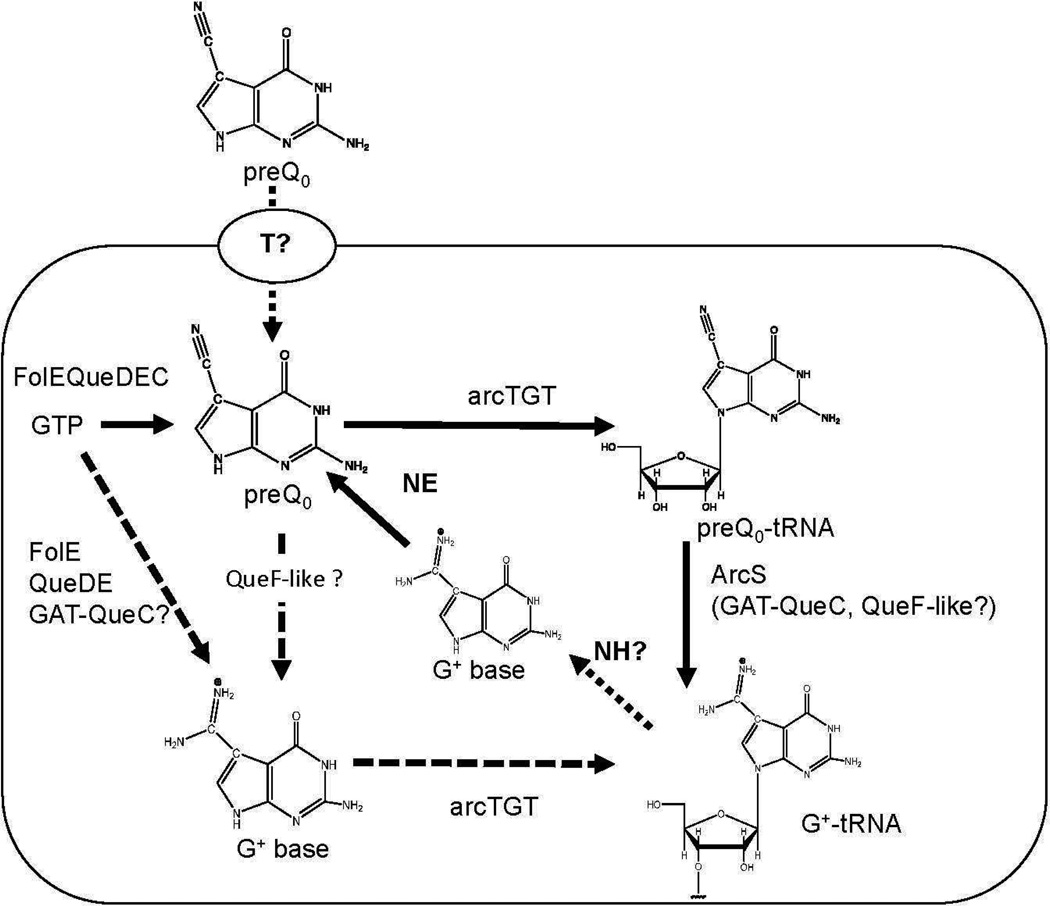
While the data clearly show that GAT-QueC and QueF-like function as amidinotransferases, generating G+ modified tRNA in E. coli (and remarkably that G+ can be tolerated in bacteria at position 34 in normally Q-containing tRNA), we do not yet know if the conversion of the nitrile to the amidino group occurs before or after preQ0 is inserted into tRNA (Figure 2D and 2E). The bacterial TGT (bTGT) can utilize preQ0 as a substrate (12, 25), and preQ0 nucleoside is indeed detected in •queF mutants (Figure 3A, Supplemental Figure 2, and (12)). Therefore it’s possible that the QAT-QueC and QueF-like enzymes modify preQ0–tRNA just as ArcS does. Consistent with this proposal is the observation that G+ base is very unstable (10, 11), readily undergoing deamination to reform preQ0 (Figure 4). However, bTGT is promiscuous (25), and should be able to use the G+ base as a substrate if it were available. Notably, while biochemical analysis of the canonical arcTGT has demonstrated that it isn’t able to utilize G+ (10, 11), structural comparison of the canonical arcTGT with 3D homology models of the catalytic domains of arcTGT enzymes from Crenarchaeota that lack ArcS (Supplemental Figure 6) reveal differences in the active site (see supplemental Fig. 6) that might allow accommodation of G+ base in the active sites of these crenarchaeal arcTGTs. Thus, at this point both preQ0 and preQ0-tRNA can be considered viable candidates as the natural substrate for the GAT-QueC and QueF-like enzymes. Differentiating between these possibilities will require detailed biochemical and enzymological characterizations of these novel amidinotransferase families and of the crenarchaeal arcTGT, and is currently being investigated.
Figure 4. Predicted G+ biosynthesis and salvage pathways in Crenarchaeota.
Abbreviations not in text: NE=Non enzymatic, T=predicted transporter. The solid black arrows denote the experimentally validated pathway. The dashed arrows show the predicted crenarchaeal pathway.
Finally, specific Archaea such as Sulfolobus tokodaii have retained ArcS in addition to GAT-QueC (Figure 1B). A salvage route to G+ is known to occur in Archaea (6). An abundant source of G+ precursor is the hydrolyzed archaeal tRNA. As the liberated G+ base will quickly deaminate to preQ0, the salvage route in Crenarchaeota could require ArcS (Figure 4). This work illustrates the power of comparative genomics approaches, particularly when combined with biochemical reasoning, in discovering novel enzymes and pathways, which has now led to a much more diverse picture of G+ synthesis in Archaea than previously appreciated.
METHODS
Bioinformatics
Analysis of the Archaeosine sub-system was performed in the SEED database (26). Results and protein sequences are available in the “Queuosine and Archaeosine biosynthesis” sub-system on the public SEED server (http://theseed.uchicago.edu/FIG/-index.cgi). The list of arcTGT and sequences used in these studies is given in Supporting Information. We used the Blast tools and resources at NCBI (27). Multiple protein alignments were performed with the ClustalW tool (28) in the SEED database or the MultiAlign software (http://omics.pnl.gov/). The 3D models were generated using the protein fold recognition protocols of Phyre (http://www.sbg.bio.ic.ac.uk/~phyre/, (29)) based on one- and three-dimensional sequence profiles, coupled with secondary structure and solvation potential information. Structure based multisequence alignment was performed using MultiProt (30) and ESPript (31) through the web interfaces (http://bioinfo3d.cs.tau.ac.il/MultiProt/ ) and (http://espript.ibcp.fr/ESPript/ESPript/ ), respectively.
Media, strain and plasmids
E.coli bulk tRNA extraction and analysis
Bulk tRNA was prepared from cells grown in LB with 0.2% arabinose, hydrolyzed and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS-MS) as described in (32). To compare tRNA concentrations, we compared the ratio of the levels of the Ψ modified base (m/z 245) in each sample by integrating the peak area from the selected ion chromatograms. The MS/MS fragmentation data was also used to confirm the presence of the nucleosides preQ0 and G+. All tRNA extractions and analysis were performed at least twice independently.
tRNAAsp purification and analysis
tRNAAsp was extracted from bulk tRNA using a biotinylated primer (5’biotin-CCCTGCGTGACAGGCAGG-3’) bound to the streptavidin sepharose resin (33). The RNase T1 digestion and oligonucleotide sequencing analysis by LC-MS/MS is described in the Supporting Information.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the National Institute of General Medical Sciences (1RC2GM092602–01 subcontract to V.d.C.-L.), the National Science Foundation (CHE0910751 to P.A.L.), and NASA (NNX07AJ26G to D. I.-R.). The authors would like to thank S. Alvarez for the LC-MS analysis of tRNA nucleosides, M. Dyal-Smith for the H. waslbyi strain and guidance on its culture conditions, S. Lesley (JCSG) for the SSO0016 expressing clone and T. Lowe (UCSC) for the P. calidifontis cell paste. MB is a recipient of a postdoctoral fellowship from Human Frontier Scientific Program (HFSP).
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Grosjean H. Nucleic Acids are not boring long polymers of only four types of nucleotides. In: Grosjean H, editor. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. Landes Bioscience; 2009. pp. 1–18. [Google Scholar]
- 2.Iwata-Reuyl D. Biosynthesis of the 7-deazaguanosine hypermodified nucleosides of transfer RNA. Bioorg Chem. 2003;31:24–43. doi: 10.1016/s0045-2068(02)00513-8. [DOI] [PubMed] [Google Scholar]
- 3.Meier F, Suter B, Grosjean H, Keith G, Kubli E. Queuosine modification of the wobble Base in tRNAHis influences 'in vivo' decoding properties. EMBO J. 1985;4:823–827. doi: 10.1002/j.1460-2075.1985.tb03704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gregson JM, Crain PF, Edmonds CG, Gupta R, Hashizume T, Phillipson DW, McCloskey JA. Structure of Archaeal transfer RNA nucleoside G*-15 (2-Amino-4,7-dihydro-4-oxo-7-β-D-ribofuranosyl-1H-pyrrolo[2,3-d]pyrimidine-5-carboximidamide (Archaeosine)) J. Biol. Chem. 1993;268:10076–10086. [PubMed] [Google Scholar]
- 5.Oliva R, Tramontano A, Cavallo L. Mg2+ binding and archaeosine modification stabilize the G15 C48 Levitt base pair in tRNAs. RNA. 2007;13:1427–1436. doi: 10.1261/rna.574407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phillips G, El Yacoubi B, Lyons B, Alvarez S, Iwata-Reuyl D, de Crécy-Lagard V. Biosynthesis of 7-deazaguanosine-modified tRNA nucleosides: a new role for GTP Cyclohydrolase I. J. Bacteriol. 2008;190:7876–7884. doi: 10.1128/JB.00874-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reader JS, Metzgar D, Schimmel P, de Crécy-Lagard V. Identification of four genes necessary for biosynthesis of the modified nucleoside queuosine. J. Biol. Chem. 2004;279:6280–6285. doi: 10.1074/jbc.M310858200. [DOI] [PubMed] [Google Scholar]
- 8.McCarty RM, Somogyi Ard, Lin G, Jacobsen NE, Bandarian V. The deazapurine biosynthetic pathway revealed: in vitro enzymatic synthesis of preQ0 from guanosine 5'-triphosphate in four steps. Biochemistry. 2009;48:3847–3852. doi: 10.1021/bi900400e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blaby IK, Phillips G, Blaby-Haas CE, Gulig KS, El Yacoubi B, de Crécy-Lagard V. Towards a systems approach in the genetic analysis of archaea: accelerating mutant construction and phenotypic analysis in Haloferax volcanii. Archaea. 2010;2010:426239. doi: 10.1155/2010/426239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watanabe M, Matsuo M, Tanaka S, Akimoto H, Asahi S, Nishimura S, Katz JR, Hashizume T, Crain PF, McCloskey JA, Okada N. Biosynthesis of archaeosine, a novel derivative of 7-deazaguanosine specific to Archaeal tRNA, proceeds via a pathway involving base replacement of the tRNA polynucleotide chain. J. Biol. Chem. 1997;272:20146–20151. doi: 10.1074/jbc.272.32.20146. [DOI] [PubMed] [Google Scholar]
- 11.Bai Y, Fox DT, Lacy JA, Van Lanen SG, Iwata-Reuyl D. Hypermodification of tRNA in Thermophilic archaea. Cloning, overexpression, and characterization of tRNA-guanine transglycosylase from Methanococcus jannaschii. J. Biol. Chem. 2000;275:28731–28738. doi: 10.1074/jbc.M002174200. [DOI] [PubMed] [Google Scholar]
- 12.Van Lanen SG, Reader JS, Swairjo MA, de Crécy-Lagard V, Lee B, Iwata-Reuyl D. From cyclohydrolase to oxidoreductase: discovery of nitrile reductase activity in a common fold. Proc. Natl. Acad. Sci. U. S. A. 2005;102:4264–4269. doi: 10.1073/pnas.0408056102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noguchi S, Nishimura Y, Hirota Y, Nishimura S. Isolation and characterization of an Escherichia coli mutant lacking tRNA-guanine transglycosylase. Function and biosynthesis of queuosine in tRNA. J. Biol. Chem. 1982;257:6544–6550. [PubMed] [Google Scholar]
- 14.Slany RK, Bosl M, Kersten H. Transfer and isomerization of the ribose moiety of AdoMet during the biosynthesis of queuosine tRNAs, a new unique reaction catalyzed by the QueA protein from Escherichia coli. Biochimie. 1994;76:389–393. doi: 10.1016/0300-9084(94)90113-9. [DOI] [PubMed] [Google Scholar]
- 15.Miles ZD, McCarty RM, Molnar G, Bandarian V. Discovery of epoxyqueuosine (oQ) reductase reveals parallels between halorespiration and tRNA modification. Proc Natl Acad Sci U S A. 2011;108:7368–7372. doi: 10.1073/pnas.1018636108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phillips G, Chikwana VM, Maxwell A, El-Yacoubi B, Swairjo MA, Iwata-Reuyl D, de Crécy-Lagard V. Discovery and characterization of an amidotransferase involved in the modification of archaeal tRNA. J. Biol. Chem. 2010;285:12706–12713. doi: 10.1074/jbc.M110.102236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edmonds CG, Crain PF, Gupta R, Hashizume T, Hocart CH, Kowalak JA, Pomerantz SC, Stetter KO, McCloskey JA. Posttranscriptional modification of tRNA in thermophilic archaea (Archaebacteria) J. Bacteriol. 1991;173:3138–3148. doi: 10.1128/jb.173.10.3138-3148.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCloskey JA, Liu X-H, Crain PF, Bruenger E, Guymon R, Hashizume T, Stetter KO. Posttranscriptional modification of transfer RNA in the submarine hyperthermophile Pyrolobus fumarii. Nucleic Acids Symp Ser (Oxf) 2000;44:267–268. doi: 10.1093/nass/44.1.267. [DOI] [PubMed] [Google Scholar]
- 19.Massière F, Badet-Denisot MA. The mechanism of glutamine-dependent amidotransferases. Cell. Mol. Life Sci. 1998;54:205–222. doi: 10.1007/s000180050145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colloc'h N, Poupon A, Mornon JP. Sequence and structural features of the T-fold, an original tunnelling building unit. Proteins. 2000;39:142–154. doi: 10.1002/(sici)1097-0134(20000501)39:2<142::aid-prot4>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 21.Kim Y, Zhou M, Moy S, Morales J, Cunningham MA, Joachimiak A. High-resolution structure of the nitrile reductase QueF combined with molecular simulations provide insight into enzyme mechanism. J. Mol. Biol. 2010;404:127–137. doi: 10.1016/j.jmb.2010.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kowalak JA, Pomerantz SC, Crain PF, McCloskey JA. A novel method for the determination of posttranscriptional modification in RNA by mass spectrometry. Nucleic Acids Res. 1993;21:4577–4585. doi: 10.1093/nar/21.19.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandal D, Kohrer C, Su D, Russell SP, Krivos K, Castleberry CM, Blum P, Limbach PA, Söll D, RajBhandary UL. Agmatidine, a modified cytidine in the anticodon of archaeal tRNA(Ile), base pairs with adenosine but not with guanosine. Proc. Natl. Acad. Sci. U S A. 2010;107:2872–2877. doi: 10.1073/pnas.0914869107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sekiya T, Mori M, Takahashi N, Nishimura S. Sequence of the distal tRNA1Asp gene and the transcription termination signal in the Escherichia coli ribosomal RNA operon rrnF(or G) Nucleic Acids Res. 1980;8:3809–3828. doi: 10.1093/nar/8.17.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoops GC, Townsend LB, Garcia GA. tRNA-guanine transglycosylase from Escherichia coli: structure- activity studies investigating the role of the aminomethyl substituent of the heterocyclic substrate PreQ1. Biochemistry. 1995;34:15381–15387. doi: 10.1021/bi00046a047. [DOI] [PubMed] [Google Scholar]
- 26.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crécy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov DA, Rückert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelley LA, Sternberg MJE. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protocols. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 30.Shatsky M, Nussinov R, Wolfson HJ. A method for simultaneous alignment of multiple protein structures. Proteins: Structure, Function, and Bioinformatics. 2004;56:143–156. doi: 10.1002/prot.10628. [DOI] [PubMed] [Google Scholar]
- 31.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 32.de Crécy-Lagard V, Brochier-Armanet C, Urbonavicius J, Fernandez B, Phillips G, Lyons B, Noma A, Alvarez S, Droogmans L, Armengaud J, Grosjean H. Biosynthesis of wyosine derivatives in tRNA: an ancient and highly diverse pathway in Archaea. Mol. Biol. Evol. 2010:2062–2077. doi: 10.1093/molbev/msq096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rinehart J, Krett B, Rubio MAT, Alfonzo JD, Söll D. Saccharomyces cerevisiae imports the cytosolic pathway for Gln-tRNA synthesis into the mitochondrion. Genes Dev. 2005;19:583–592. doi: 10.1101/gad.1269305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.