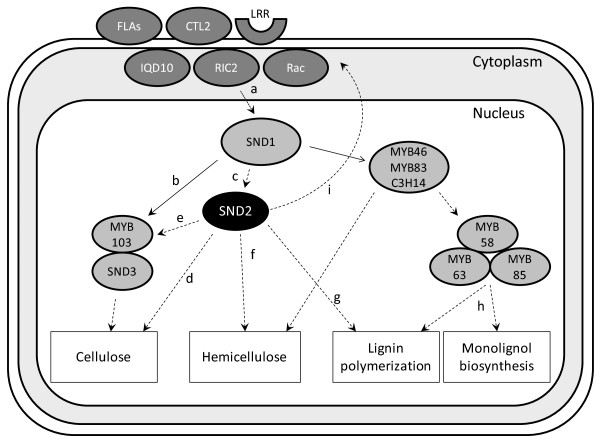
Figure 5.
Proposed model of SND2-mediated SCW regulation in IFs. Solid lines indicate known direct protein-DNA interactions. Dashed lines indicate direct or indirect protein-DNA interactions. Master regulator SND1 is activated by a signal transduction pathway proposed by Oikawa et al. [56] (a). SND1 directly activates transcription of MYB103 and SND3 (b), and indirectly activates SND2 through an unknown intermediate (c; [21]). SND2 activates cellulose-synthesizing CesAs, either directly (d) or through the activation of MYB103 (e), which is known to activate SCW cellulose gene, CesA8 [21]. SND2 regulates hemicellulosic genes (f; Table 1), independently to a similar role played by direct SND1 targets MYB46, MYB83 or C3H14 [76-78]. SND2 plays a role in lignification through activation of lignin polymerization genes LAC4 and LAC17 (g; Table 1), but it does not regulate monolignol biosynthetic genes as is the case for MYB58, MYB63 and MYB85 (h) [21,60]. SND2 activates transcription of GPI-anchored FLA11/FLA12, CTL2 and other components of the signal transduction pathway (i), which leads to upregulation of SND1 (a).