Abstract
Interleukin-17 cytokines are a family of pro-inflammatory cytokines. Our current studies found: i) IL-17 cytokines are not ubiquitously expressed, but several receptors and TRAF3IP2 are ubiquitously expressed in tissues with a few exceptions; ii) heart and vascular tissue are in the second tier of readiness to respond to IL-17 cytokine stimulation; iii) alternative transcription starting sites and alternative spliced isoforms are found in IL-17 cytokine and receptor transcripts; iv) higher hypomethylation status is associated with higher expressions of IL-17 receptors; v) the binding sites of several RNA binding proteins are found in the 3′UTRs of the mRNAs of IL-17 cytokines and receptors; and vi) numerous microRNA binding sites are statistically equivalent to that of experimentally verified microRNAs-mRNA interactions in the 3′UTRs of IL-17 cytokine and receptor mRNAs. These results suggest that mechanisms including alternative promoters, alternative splicing, RNA binding proteins, and microRNAs regulate the structures and expressions of IL-17 cytokines and receptors. These results provide an insight into the roles of IL-17 in mediating inflammation and immunity.
Keywords: interleukin-17 Cytokines, Interleukin-17 Receptors, Gene Expression, mRNA Stability, Vascular Inflammation, Review
2. INTRODUCTION
Cardiovascular disease (CVD) remains a leading cause of fatality in well-developed countries. Despite a long held understanding and strong characterization of the traditional and non-traditional risk factors for CVD, some mechanisms of CVD onset have only recently been elucidated. As a chronic inflammatory disease, atherosclerosis and its progression involve both the adaptive and innate immune systems (1). For example, we and others reported that CD4+CD25high regulatory T cells (2–4) (a type of adaptive immune cells) and Ly6Cmid/high monocytes (a type of innate immune cells) play suppressive and promoting roles respectively, in the pathogenesis of atherosclerosis and vascular inflammation (5). Since 2003, a new lineage of CD4+RORgammat+ (retinoid-related orphan receptor gamma) T cells has been defined by its production of pro-inflammatory cytokine interleukin-17 (IL-17) and hence named T-helper 17 (Th17) cells. These cells have been found to play an essential role in promoting autoimmune diseases, inflammation (6, 7), and potentially atherosclerosis (8–11) (see our invited review (12)). Increased Th17, rather than Th1 response is associated with some vasculopathy (13), suggesting an important role for IL-17 cytokines in cardiovascular diseases. However, detailed tissue expression and regulation mechanisms of IL-17 cytokines and receptors remain poorly defined in cardiovascular and other tissues.
IL-17 cytokine family consists of six members designated IL-17A-F. IL-17 cytokine family members are highly conserved between human and mouse with homology of 62% to 88%(14) among the six members across species. The first cytokine of this family studied was IL-17A (also known as IL-17)(15, 16). Sequentially, the other five members were cloned and categorized into the IL-17 cytokine family (17–20). IL-17 cytokines are a family of pro-inflammatory cytokines implicated in numerous autoimmune and inflammatory diseases. IL-17 cytokines have found to induce expression of pro-inflammatory cytokines and chemokines including CXCL1, CXCL2, IL-6, and G-CSF(21, 22) via the activation of nuclear factor - kappaB (NF-κB) and mitogen-activated protein (MAP) kinase pathways (23, 24). It has been shown that IL-17 and IL-17F have the ability to form dimers, and the main functions of IL-17, IL-17F, and IL-17A/F are in autoimmune pathology, extracellular pathogen immunity, and neutrophil recruitment (7, 25). However, the issue of which tissues express both dimer-forming cytokines remains poorly defined.
The IL-17 receptor family is made up of five unique cytokine receptor members: IL-17RA, IL-17RB, IL-17RC, IL-17RD, and IL-17RE(26). Similar to the cytokines, IL-17 receptors between human and mouse also show a great degree of homology (68% to 90%)(14). IL-17RA and IL-17RC are the receptors for IL-17 and IL-17F, and they are the best characterized receptors among this family of cytokine receptors (27–29). IL-17RB has affinity for IL-17B and IL-17E, IL-17RE binds IL-17C, and the ligands for IL-17RD remain to be elucidated (18, 30). IL-17RA has been shown to be ubiquitously expressed but at higher levels in hematopoietic tissues (27). IL-17 has been shown to activate some of its pro-inflammatory effects via NF-κB and MAPK pathways. A component of the IL-17 receptors that is crucial for IL-17 signaling is an adaptor termed ACT1 (also known as TRAF3IP2; TRAF3 interacting protein 2), which is needed to mediate various downstream event of the IL-17 receptors (31–34). However, the issue of which tissues express receptor complexes remains poorly defined.
Despite significant progress, several important knowledge gaps exist which prevent investigators from defining the detailed roles of these molecules in inflammation and immune responses. First, how IL-17 cytokines and receptors are expressed in cardiovascular and other tissues; second, whether alternative splicing and alternative promoters regulate the structures of IL-17 cytokines and receptors; and third, whether mRNA decay proteins (AU-rich element binding proteins)(35, 36) and microRNAs (37) regulate the mRNA stability and translation of IL-17 cytokines and receptors. Using database mining techniques and statistical analysis similar to that we reported previously (38, 39), we examined the expressions of IL-17 cytokines and receptors in cardiovascular and other tissues from a panoramic viewpoint. In addition, we also examined the potential molecular mechanisms regulating the expression of these IL-17 cytokines and receptors. In depth analysis of the expression patterns of these important cytokines and receptors could prove vital in further understanding the underlying mechanism for immune responses and inflammation. This insight may provide novel avenues for innovative therapeutic treatments for pro-atherogenic inflammation and other cardiovascular diseases.
3. METHODS
3.1. Tissue expression profiles of genes encoding IL-17 cytokines, IL-17 receptors, RORC, and TRAF3IP2
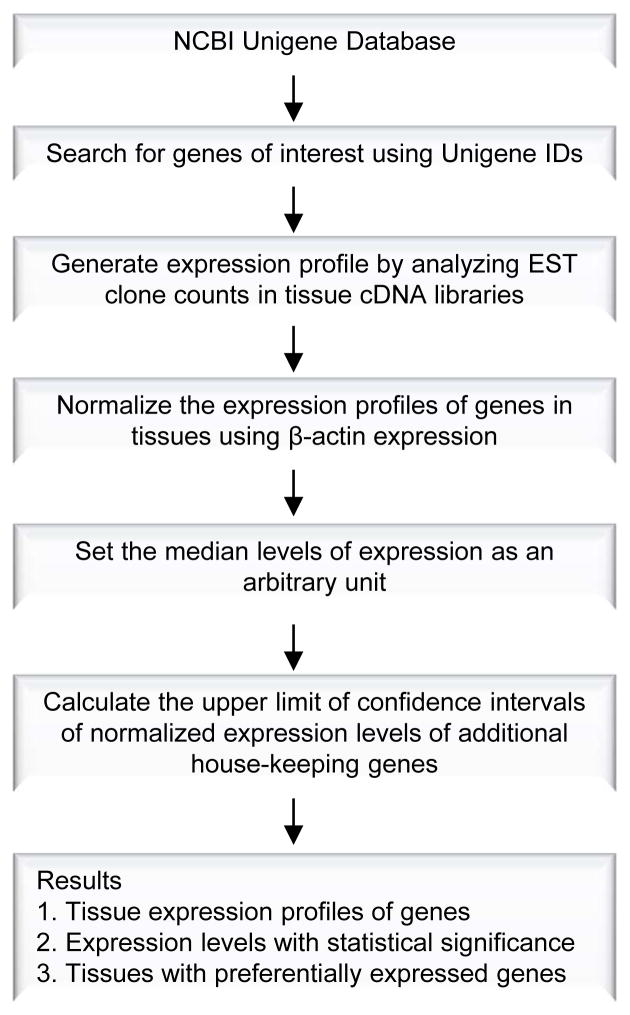
Experimental data mining strategy, as we previously described (38–40) (Figure 1), was used to analyze the expression profiles of mRNA transcripts of IL-17 cytokines, IL-17 receptors, RORC, and TRAF3IP2 in cardiovascular and other tissues in humans and mice by mining experimentally verified human and mouse mRNA transcript expressions in the sequence tag (EST) databases of the National Institutes of Health (NIH)/National Center of Biotechnology Information (NCBI) Unigene (http://www.ncbi.nlm.nih.gov/sites/entrez?db=unigene)(41). Transcripts per million of genes of interest were normalized with that of house-keeping beta-actin in any given tissue to calculate the arbitrary units of the gene expression. The confidence interval of the expression variation of house-keeping genes was generated by calculating the mean and 2 times the standard deviation of the arbitrary units of three randomly selected house-keeping genes (RPS27A, GADPH, and ARHGDIA in human; Ldha, Nono, and Rpl32 in mouse) normalized by beta-actin in given tissues. If the expression variation of a given gene in the tissues was larger than the upper limit of the confidence interval (the mean plus 2 times the standard deviation) of the housekeeping genes, the high expression levels of genes in the tissues were statistically significant. Any given gene transcript, if lower than one per million, was technically presented as no expression.
Figure 1.
Flow Chart of Database Mining Analysis of Gene Expression Profiles Using NCBI/UniGene Database.
3.2. Alternative spliced isoforms of IL-17 cytokines and IL-17 receptors
The presence and features of alternative promoters and alternatively spliced isoforms of each gene were examined with the AceView database of the National Institutes of Health (NIH)/National Center of Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/index.html).
3.3. Presence of AU-rich elements and functional motifs in 3′ untranslated regions of IL-17 cytokines and IL-17 receptors
The gene of interest was searched in the UTRdb (http://utrdb.ba.itb.cnr.it/search) at the Institute for Biomedical Technologies, University of Bari for the existence of functional motifs and signals in the 3′ untranslated regions (3′UTR) of each mRNA. The presence of AU-rich elements in the 3′UTRs of IL-17 cytokines and receptors was searched using the AU-rich Element Containing mRNA Database (ARED 3.0) (http://brp.kfshrc.edu.sa/ARED/). AU-rich elements or AREs have been classified into three classes based on the presence and distribution of AUUUA sequence (42).
3.4. Correlation of the ratios of tissue SAH versus SAM concentrations with the expression levels of IL-17 cytokine and receptor mRNA transcripts
The concentrations of S-adenosylmethionine (SAM) levels over S-adenosylhomocysteine (SAH) levels were determined previously by Ueland’s group (43–45) in tissues from adult male mice under physiological condition. The SAM and SAH levels were measured in perchloric acid extracts by high performance liquid chromatography. The SAM/SAH ratios were calculated for the current analysis based on Ueland’s results (43–45). Tissue SAH and SAM concentrations and SAM/SAH ratios were used for further comparison and regression analyses. Simple linear regression analyses were performed using Sigma Plot 9.0 (Systat Software, Inc, San Jose, CA, USA) by plotting mRNA levels of individual gene against SAM/SAH ratios in seven mouse tissues including the brain, kidney, liver, spleen, heart, lung, and thymus. Multivariable regression analyses were performed to evaluate the effect of SAM/SAH ratios on the expressions of IL-17 cytokines and receptors.
3.5. MicroRNA interaction with the mRNAs of IL-17 cytokines and IL-17 receptors
The interactions of the mRNAs of genes of interest with microRNAs were examined using the Bioinformatics and Research Computing software TargetScan Release 5.1. (http://www.targetscan.org/) from the Whitehead Institute for Biomedical Research of Massachusetts Institute of Technology (MIT). The significance of microRNAs binding to the genes of interest was determined using the confidence intervals generated from the microRNAs within the Tarbase, an experimentally verified microRNA online database (http://diana.cslab.ece.ntua.gr/tarbase/)(46, 47). Briefly, human microRNAs, which were single site effective and confirmed with luciferase reporter assays, were used for establishing the intervals. Using single site effective and luciferase assay confirmed microRNAs ensures that the interactions between the microRNAs and their respective mRNA targets are specific. 27 microRNAs that met the criteria were selected and evaluated in TargetScan to construct the intervals and set the lower limit for the context values and score percentile. MicroRNAs with the context score of 70% or higher and context value of -0.22 or lower were determined to be significant.
4. RESULTS
4.1. Most of IL-17 cytokines are not constitutively expressed in the tissues examined, but several IL-17 receptors and TRAF3IP2 are ubiquitously expressed
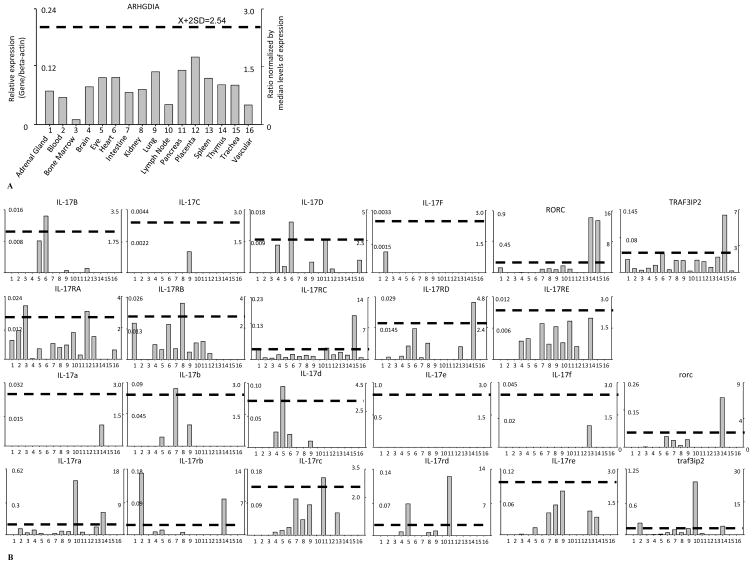
We hypothesized that in order to keep inflammation in check, IL-17 cytokines and receptors are differentially expressed in cardiovascular and other tissues. To examine this hypothesis, a database mining method as we reported (40) was used to examined experimentally verified expression profiles of mRNA transcripts of IL-17 cytokines and receptors (Table 1). The copy number per million transcripts was calculated based on the experimental data of expression sequence tag (EST) cDNA cloning and sequencing in the NCBI UniGene database. The gene expression data were normalized by beta-actin expression data in the same tissue, thus the arbitrary units of gene expression were comparable among genes (Figure 2). Statistical significance is defined when gene expression is larger than the upper limit of the confidence interval (the mean plus 2 times the standard deviation) of the housekeeping genes. As shown in Table 2A and 2B, the expressions of six IL-17 cytokines, IL-17A-F, five IL-17 receptors, IL-17RA-E, RORC, and TRAF3IP2 (ACT1) gene transcripts were examined in 16 tissues: adrenal gland, blood, bone marrow, brain, eye, heart, intestine, kidney, lung, lymph node, pancreas, placenta, spleen, thymus, trachea, and vascular. The EST profiles for human IL-17A, human IL-17E, mouse IL-17c, and mouse IL-17e were not found in the EST NCBI UniGene database, suggesting lower abundance of these molecules than other IL-17 cytokines expressed in tissues. Of the 16 tissues examined, only a few tissues expressed IL-17 cytokines, which correlate with previous reports (15). But our analysis examined more cardiovascular inflammation-related human and mouse tissues for expression of IL-17 cytokines than previous reported. These expression patterns suggest that the non-detected IL-17 cytokines are not required for physiological functions of these tissues; and that the expressions of these non-detected IL-17 cytokines are not beneficial for physiological functions of these tissues.
Table 1.
The Unigene id of human and mouse genes examined
| Genes | Unigene ID | Genes | Unigene ID |
|---|---|---|---|
| L-17B | Hs. 156979 | IL-17a | Mm.5419 |
| IL-17C | Hs.278911 | IL-17b | Mm.59313 |
| IL-17D | Hs.655142 | IL-17d | Mm.390726 |
| IL-17F | Hs.272295 | IL-17e | Mm.90154 |
| RORC | Hs.256022 | IL-17f | Mm.222807 |
| IL-17RA | Hs.129751 | rorc | Mm. 4372 |
| IL-17RB | Hs.654970 | IL-17ra | Mm.4481 |
| IL-17RC | Hs.129959 | IL-17rb | Mm.269363 |
| IL-17RD | Hs.150725 | IL-17rc | Mm.213397 |
| IL-17RE | Hs.390823 | IL-17rd | Mm.206726 |
| TRAF3IP2 | Hs.561514 | IL-17re | Mm.131781 |
| traf3ip2 | Mm.436686 |
Figure 2.
A. Data Presentation Format (The data presented in X-, Y-axis and tissue order are applied to all the genes examined). B. Tissue Expression Profiles of IL-17 cytokines and receptors, RORC, and TRAF3IP2. The gene expression profiles of IL-17 cytokines and receptors, TRAF3IP2, and RORC in mouse and human tissues. A) As an example, the gene expression profiles of human housekeeping gene Rho GDP dissociation inhibitor (GDI) alpha (ARHGDIA) in the sixteen tissues including adrenal gland, blood, bone marrow, brain, eye, heart, intestine, kidney, lung, lymph node, pancreas, placenta, spleen, thymus, trachea, and vascular are presented, with the tissue names and position numbers are shown on the X-axis. The gene expression data were normalized by the β-actin (Hs.520640) expression data from the same tissue, which are presented on the left Y-axis. The expression ratios among tissues were generated by normalizing the arbitrary units of the gene in the tissues with the median level of the arbitrary units of the gene in all the tissues which are presented on the right Y-axis. In order to define confidence intervals for statistically higher expression levels of given genes, we calculated the confidence intervals of tissue expression [the mean X + 2 x standard deviations (SD) = 2.54] for three housekeeping genes including Rho GDP dissociation inhibitor alpha (ARHGDIA, Hs.159161), glyceraldehyde-3-phosphate dehydrogenase (GAPDH, Hs.544577), and ribosomal protein S27a (RPS27A, Hs.311640). The expression variations of given genes in tissues, when they were larger than 2.54-fold, were defined as the high expression levels with statistical significance (the right Y-axis). To define confidence intervals for statistically higher expression levels of given genes in mouse tissue, we calculated the confidence intervals of tissue expression [the mean X + 2 x standard deviations (SD) = 2.67] for three mouse housekeeping genes including Lactate dehydrogenase A (Ldha, Mm.29324), non-POU-domain-containing octamer binding protein (Nono, Mm.280069), and ribosomal protein L32 (Rpl32, Mm.104368). The expression variations of given genes in tissues, when they were larger than 2.67-fold, were defined as the high expression levels with statistical significance (the right Y-axis). B) The expression profiles of IL-17 cytokines and receptors, TRAF3IP2, and RORC in human tissues (top two rows, with cytokine and receptor family members designated with capital letters) and mouse tissues (bottom two rows, with cytokine and receptor family members designated with lowercase letters). The X-axis indicates the sixteen tissues examined in the same order as that shown in Figure 2A with position numbers shown.
Table 2.
IL-17 cytokines and receptors are differentially expressed in human and mouse tissues
| A. IL-17 cytokines and receptors are differentially expressed in human tissues | |||
|---|---|---|---|
| Tissues | Cytokine and RORC | Tissues | Receptor and TRAF3IP2 |
| 1. Adrenal Gland | RORC | 1. Adrenal Gland | IL-17RA, IL-17RB, IL-17RC, TRAF3IP2 |
| 2. Blood | IL-17F | 2. Blood | IL-17RA, IL-17RC, IL-17RD, TRAF3IP2 |
| 3. Bone Marrow | 3. Bone Marrow | IL-17RA, IL-17RC, TRAF3IP2 | |
| 4. Brain | IL-17D, RORC | 4. Brain | IL-17RA, IL-17RB, IL-17RC, IL-17RD, IL-17RE, TRAF3IP2 |
| 5. Eye | IL-17B, IL-17D | 5. Eye | IL-17RA, IL-17RB, IL-17RC, IL-17RD, IL-17RE, TRAF3IP2 |
| 6. Heart | IL-17B, IL-17D | 6. Heart | IL-17RB, IL-17RC, IL-17RD, TRAF3IP2 |
| 7. Intestine | RORC | 7. Intestine | IL-17RA, IL-17RB, IL-17RC, IL-17RD, IL-17RE, TRAF3IP2 |
| 8. Kidney | RORC | 8. Kidney | IL-17RA, IL-17RB, IL-17RC, IL-17RD, IL-17RE, TRAF3IP2 |
| 9. Lung | IL-17B, IL-17C, IL-17D, RORC | 9. Lung | IL-17RA, IL-17RB, IL-17RC, IL-17RE, TRAF3IP2 |
| 10. Lymph Node | RORC | 10. Lymph Node | IL-17RA, IL-17RB, IL-17RE, TRAF3IP2 |
| 11. Pancreas | IL-17D, RORC | 11. Pancreas | IL-17RA, IL-17RB, IL-17RC, IL-17RE, TRAF3IP2 |
| 12. Placenta | IL-17B, IL-17D | 12. Placenta | IL-17RA, IL-17RB, IL-17RC, IL-17RE, TRAF3IP2 |
| 13. Spleen | 13. Spleen | IL-17RA, IL-17RC, IL-17RD, TRAF3IP2 | |
| 14. Thymus | RORC | 14. Thymus | IL-17RC, IL-17RE, TRAF3IP2 |
| 15. Trachea | RORC | 15. Trachea | IL-17RC, IL-17RD, TRAF3IP2 |
| 16. Vascular | IL-17D | 16. Vascular | IL-17RA, IL-17RC, TRAF3IP2 |
| B. IL-17 cytokines and receptors are differentially expressed in mouse tissues | |||
| Tissues | Cytokine and RORC | Tissues | Receptor and TRAF3IP2 |
| 1. Adrenal Gland | 1. Adrenal Gland | ||
| 2. Blood | 2. Blood | IL-17ra, IL-17rb, traf3ip2 | |
| 3. Bone Marrow | rorc | 3. Bone Marrow | IL-17ra, IL-17re, traf3ip2 |
| 4. Brain | IL-17d | 4. Brain | IL-17ra, IL-17rb, IL-17rc, IL-17rd, traf3ip2 |
| 5. Eye | IL-17b, IL-17d | 5. Eye | IL-17ra, IL-17rb, IL-17rc, IL-17rd, IL-17re, traf3ip2 |
| 6. Heart | IL-17d, rorc | 6. Heart | IL-17rc, traf3ip2 |
| 7. Intestine | IL-17b, rorc | 7. Intestine | IL-17ra, IL-17rc, IL-17re, traf3ip2 |
| 8. Kidney | rorc | 8. Kidney | IL-17ra, IL-17rb, IL-17rc, IL-17rd, IL-17re, traf3ip2 |
| 9. Lung | IL-17b, IL-17d, rorc | 9. Lung | IL-17ra, IL-17rc, IL-17rd, IL-17re, traf3ip2 |
| 10. Lymph Node | 10. Lymph Node | IL-17ra, traf3ip2 | |
| 11. Pancreas | 11. Pancreas | IL-17ra, IL-17rc, IL-17rd, traf3ip2 | |
| 12. Placenta | 12. Placenta | ||
| 13. Spleen | IL-17f | 13. Spleen | IL-17ra, IL-17rc, IL-17re |
| 14. Thymus | IL-17a, rorc | 14. Thymus | IL-17ra, IL-17rb, IL-17re, traf3ip2 |
| 15. Trachea | 15. Trachea | ||
| 16. Vascular | 16. Vascular | ||
Among the IL-17 cytokines that had expression (Table 2A), human IL-17D was most widely expressed, and it was found in vascular, placenta, heart, brain, pancreas, eye, and lung. IL-17D has been shown to have an important role in endothelial cell pathology; in HUVECs (Human Umbilical Vein Endothelial Cells) IL-17D upregulates pro-inflammatory cytokines IL-6 and IL-8 production and induces GM-CSF(48). Human IL-17B was expressed in placenta, heart, eye, and lung. IL-17B and IL-17D were expressed significantly in the heart. Human IL-17F and IL-17C were expressed in blood and lung, respectively. In addition, we found that Th17 cell-specific transcription factor RORC (RORgamma) was expressed in trachea, adrenal gland, brain, pancreas, thymus, kidney, lung, and intestine. The tissue expression pattern of RORC was broader than that of Th17 cytokines IL-17A and IL-17F, which was conserved evolutionally in mouse and humans. These results suggest that other factors may also be involved in the development of IL-17A- and IL-17F-secreting Th17 cells. Mouse IL-17 cytokines expressed in tissues differently from that in human tissues but the mouse tissue expression of IL-17d was similar to that in human tissues (Table 2B). In mouse, expressions of IL-17b in intestine and IL-17d in eye were statistically significant.
IL-17 receptor A (IL-17RA), IL-17RC, and TRAF3IP2 were expressed in all human tissues examined except in trachea, heart, and thymus for IL-17RA, and in lymph nodes for IL-17RC, which correlate with others findings that IL-17RA was found in human epithelial cells, fibroblast, B and T lymphocytes, stromal cells (49), and vascular endothelial cells (26). IL-17RA expressions in human bone marrow and placenta were statistically significant. IL-17RB in human kidney was expressed significantly. IL-17RC expressions in human pancreas and trachea were statistically significant. In addition, IL-17RB was expressed in 10 out of 16 tissues; IL-17E was also expressed in 9 out of 16 tissues; IL-17RD was expressed in 8 out of 16 tissues. The tissue expression patterns of IL-17 receptors and TRAF3IP2 were most conserved among mouse and human tissues. Also, in mouse statistically significant expression were found in lymph node and thymus for IL-17ra, in blood and thymus for IL-17rb, in pancreas for IL-17rc, and in pancreas and eye for IL-17rd. Expression of TRAF3IP2 was significant in human trachea while the expression of this gene in mouse was found to be significant in blood, lymph node, thymus, and lung.
4.2. Heart and vascular tissues are in the second tier of readiness to respond to IL-17 cytokine stimulation, which requires the upregulation of IL-17 receptor complex components
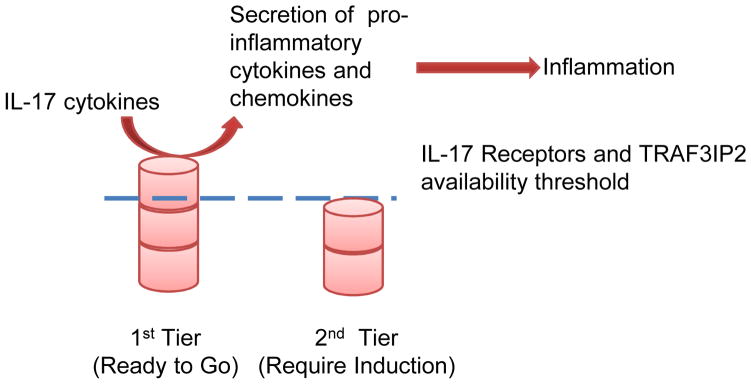
IL-17 and IL-17F define a new lineage of IL-17-producing CD4+ T helper (Th17) cells (50, 51). We hypothesized that functional status of IL-17 receptor complex can be different among tissues based on the expression status of IL-17 receptor complex and TRAF3IP2. Since the complex of IL-17RA and IL-17RC is required for IL-17 and IL-17F signaling (29), and TRAF3IP2 is essential for IL-17 signaling (52, 53), we divided the tissues examined into two tiers based on their expression of IL-17 receptor complexes and TRAF3IP2 (Table 3). IL-17RA/B is required for IL-17E signaling (54, 55). Tissues that express IL-17 receptor complexes (A/C or A/B) and TRAF3IP2 are placed in the first tier of “ready to go” status. Tissues that do not express TRAF3IP2 or all the receptors are placed in the second tier of “inducible” status with induction/upregulation of one or more components needed for a complete complex of IL-17 signaling (Figure 3).
Table 3.
The two-tier expression status of IL-17 cytokine receptors are identified in human and mouse tissues
| IL-17 Receptor Complex | Human Tissue | Mouse Tissue |
|---|---|---|
| First tier (“ready to go” expression status with all components) | ||
| IL-17RA/C Complex +TRAF3IP2 | Blood, vascular, placenta, bone marrow, adrenal gland, brain, pancreas, eye, spleen, kidney, lung, intestine | Brain, pancreas, eye, spleen, kidney, lung, intestine |
| IL-17RA/B Complex +TRAF3IP2 | Placenta, adrenal gland, brain, lymph node, pancreas, eye, kidney, lung, intestine | Blood, brain, eye, thymus, kidney |
| Second tier (“inducible” expression status that requires up-regulation of at least one component) | ||
| IL-17RA/C Complex +TRAF3IP2 | Trachea, heart, lymph node, thymus | Blood, vascular, placenta, trachea, bone marrow, adrenal gland, heart, lymph node, thymus |
| IL-17RA/B Complex +TRAF3IP2 | Vascular, trachea, blood, bone marrow, heart, spleen, thymus | Vascular, placenta, trachea, bone marrow, adrenal gland, heart, lymph nodes, pancreas, spleen, lung, intestine |
Tissues examined are categorized into first tier or second tier of readiness to respond to IL-17 stimulation based on their expression of IL-17 receptor complexes and TRAF3IP2. Tissues in the first tier of readiness express both IL-17 RA/C or IL-17RA/B and TRAF3IP2 are in a “ready to go” status. Tissues in the second tier of expression are in an “inducible” status which requires up-regulation of at least one component for IL-17 signaling.
Figure 3.
The Two-Tier Model of IL-17 Receptors and TRAF3IP2 Expression. in Human and Mouse Tissues. Tissues in the first tier expression status will secrete pro-inflammatory cytokines and chemokines to induce inflammation in response to IL-17 cytokine stimulation while tissues in the second tier need up-regulation of at least one component in the IL-17 receptor complex to induce secretion of pro-inflammatory cytokines and chemokines to drive the inflammatory process.
Of note, in term of expressions of both IL-17RA/C complex and IL-17RA/B complex, heart was in the second tier. IL-17 signaling may participate in chronic inflammation in heart in response to stimulation by pro-inflammatory risk factors. For the expression of IL-17RA/B complex, vascular tissue was also in the second tier, suggesting that IL-17RA/B complex may be involved in acute and chronic inflammation in vascular tissue, and that IL-17RA/C complex and IL-17RA/B complex may participate in chronic inflammation in heart in response to pro-inflammatory stimuli of risk factors. Moreover, more human tissues were in the first tier than mouse tissues, suggesting that the complexes of IL-17RA/C and IL-17RA/B are more involved in the inflammation of these human tissues than in those tissues of mouse. In the most of the first tier tissues in human including placenta, adrenal gland, brain, pancreas, eye, kidney, lung, and intestine, the expressions of IL-17RA/C and IL-17RA/B signaling pathways were overlapped, which may suggest the assurance of function of IL-17 signaling in these tissues.
4.3. Alternative promoter and alternative splicing regulate the structures and expressions of IL-17 cytokines and receptors
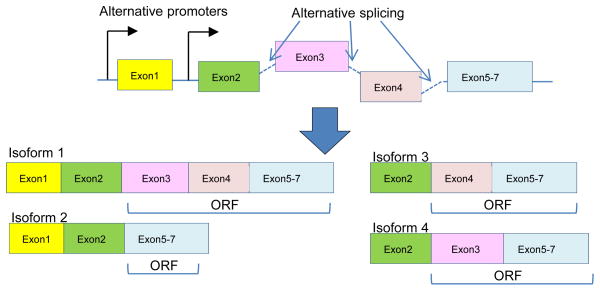
Recent findings justify a renewed interest in alternative splicing, which affects the expression of 60% of human genes. Alternative splicing explains how a vast mammalian proteomic complexity is achieved with a limited number of genes (56). Alternative splicing regulation not only depends on the interaction of splicing factors with splicing enhancers and silencers in the pre-mRNA, but also on the coupling between transcription and splicing. This coupling is possible because splicing is often co-transcriptional, and promoter identity and occupation may affect alternative splicing (57). Due to the lack of expression data for each of the alternatively spliced isoforms of IL-17 cytokines and IL-17 receptors in the database, we focused on the roles of alternative splicing in regulating the structure of IL-17 cytokines and IL-17 receptors. We hypothesized that alternative promoter and alternative splicing regulate the structure of IL-17 cytokines and the receptors. To test this hypothesis, we examined the AceView-NCBI database, the NIH-supported, most comprehensive database of alternative promoters and alternatively spliced isoforms of genes based on experimental data of cDNA cloning and DNA sequencing analysis of tissue mRNA transcriptomes (58). As shown in Table 4A and 4B, human IL-17 and mouse IL-17 had no alternatively spliced isoform, suggesting the functional intolerance for structural variations of these molecules. In contrast, other human IL-17 cytokines and mouse IL-17 cytokines had numerous isoforms. Human IL-17B, IL-17D, and IL-17F had alternative promoters, suggesting that the importance of alternative promoters of IL-17 cytokines in response to tissue-specific and/or stimulation-specific transcriptional regulation (Figure 4). Human IL-17RA (18 isoforms), IL-17RC (26 isoforms), and IL-17RD (4 isoforms) had more than three open reading frames (ORFs). These results suggest that alternative splicing plays more important roles in regulating the structures of IL-17 receptors than regulating the structures of IL-17 cytokines, which may serve as a compensatory mechanism since IL-17 receptors were more widely expressed than IL-17 cytokines in the tissues examined. Future work is needed to determine whether pro-atherogenic risk factors regulate alternative splicing and alternative promoters.
Table 4.
Alternative promoter and alternative splicing regulate the expression and structures of IL-17 cytokines and receptors
| A. Alternative promoter and alternative splicing regulate the expression and structures of IL-17 Cytokines | ||||||
|---|---|---|---|---|---|---|
| Gene | Exon(s) | Total Isoform(s) | ORF Isoform(s) | IL-17 domain /secreted | Promoter(s) | |
| IL-17A | 3 | 1 | 1 | 1 | 1 | |
| IL-17B | 6 | 3 | 3 | 2 | 2 | |
| IL-17C | 4 | 2 | 2 | 1 | 1 | |
| IL-17D | 8 | 5 | 5 | 2 | 3 | |
| IL-17E | 3 | 2 | 2 | 2 | 1 | |
| IL-17F | 3 | 2 | 2 | 1 | 2 | |
| IL-17a | 3 | 1 | 1 | 1 | 1 | |
| IL-17b | 8 | 7 | 7 | 4 | 4 | |
| IL-17c | 2 | 1 | 1 | 1 | 1 | |
| IL-17d | 3 | 3 | 2 | 2 | 1 | |
| IL-17e | 3 | 1 | 1 | 1 | 1 | |
| IL-17f | 9 | 4 | 4 | 2 | 2 | |
| B. Alternative promoter and alternative splicing regulate the expression and structures of IL-17 receptors | ||||||
| Gene | Exon(s) | Total Isoform(s) | ORF Isoform(s) | Secreted | SEFIR domain | Promoter(s) |
| IL-17RA | 33 | 23 | 18 | <18 | 3 | |
| IL-17RB | 11 | 2 | 1 | 1 | 11 | |
| IL-17RC | 39 | 28 | 26 | 5 | <26 | 4 |
| IL-17RD | 18 | 5 | 4 | <4 | 1 | |
| IL-17RE | 13 | 1 | 1 | 1 | 11 | |
| IL-17ra | 14 | 4 | 3 | 1 | <3 | 1 |
| IL-17rb | 11 | 1 | 1 | 1 | 11 | |
| IL-17rc | 20 | 7 | 7 | <7 | 2 | |
| IL-17rd | 12 | 1 | 1 | 1 | 11 | |
| IL-17re | 23 | 11 | 11 | 3 | <11 | 2 |
Of note: The data were retrieved from the NIH-NCBI-AceView database except those marked with 1, which were retrieved from the NIH-NCBI-Gene database. AceView-NCBI database was used to examine alternative promoter and alternative spliced isoforms of genes. A majority of the IL-17 cytokine and receptor genes in both human and mouse has numerous isoforms. Many of the genes have alternative promoters and more than one open reading frame (ORF) suggesting that alternative promoters and alternative splicing regulate the structure and expression of IL-17 cytokines and receptors. Human cytokine and receptor family members are designated with capital letters while mouse cytokine and receptor family members are denoted with lowercase letters.
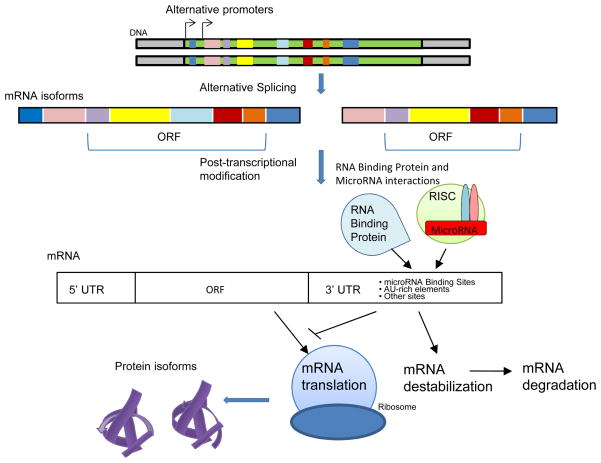
Figure 4.
Schematic Representation of Alternative Promoter and Alternative Splicing Mechanism in Generating Different Isoforms of IL-17 Cytokines and Receptors. Alternative promoter and alternative splicing regulate the expression and structure of IL-17 cytokines and receptors. Schematic representation of how alternative promoter and alternative splicing affect the open reading frame of a gene and contribute to transcription of various isoforms of the gene. In the 3′UTR of gene there is also possible modification by alternative splicing.
4.4. Higher hypomethylation status is positively associated with higher expressions of IL-17 receptors and lower expression of IL-17d in mouse tissues
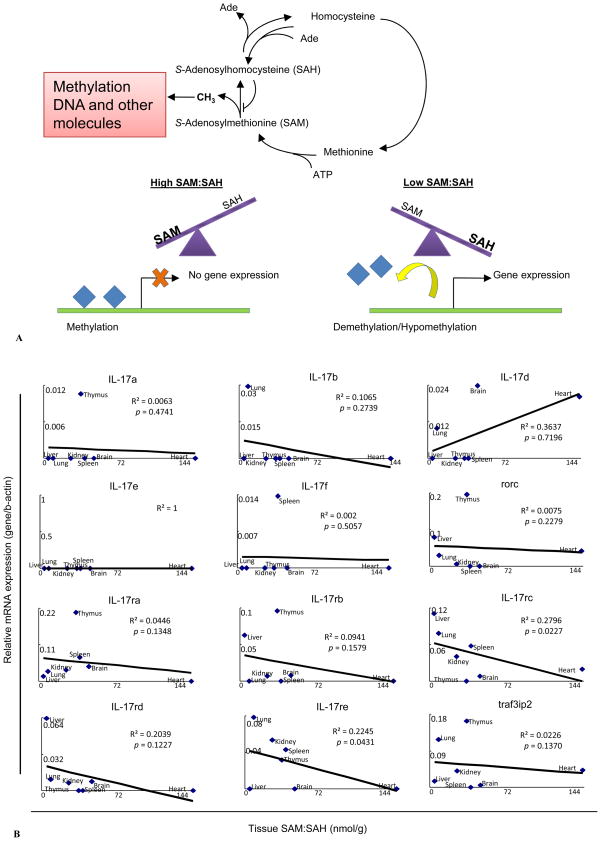
Previously, it has been shown that epigenetic changes at the IL-17A/F locus are associated with Th17 differentiation (59). To demonstrate the possibility that the expressions of IL-17 cytokines and receptors are regulated by intracellular metabolic stimuli, we hypothesized that the expressions of IL-17 cytokines and receptors are under the regulation of methylation/demethylation, a major metabolic stress-related epigenetic modification (60). As we discussed in our invited review (61), the ratio of S-adenosylmethionine (SAM) levels over S-adenosylhomocysteine (SAH) levels is an important metabolic indicator of cellular methylation status (Figure 5A)(62, 63). To test this hypothesis, we summarized tissue concentrations of SAH and SAM and the ratio of SAM over SAH in seven mouse tissues including brain, heart, kidney, liver, lung, and thymus (39), which were reported previously by Ueland’s group (43–45) (Table 5). In this study, we used SAH and SAM data generated by this group for the consideration of methodology consistency as we reported (39). We performed multivariable regression analyses to determine the effect of cellular methylation indicated by the SAM/SAH ratio on the expressions of IL-17 cytokines and receptors. As shown in Figure 5B, the SAM/SAH ratios negatively correlated with the expression levels of five IL-17 receptors and traf3ip2, especially with those of IL-17rc and IL-17re (p<0.05). The SAM/SAH ratios did not correlate with the expressions of cytokines IL-17a, IL-17e (IL-25), IL-17f, and RORgamma (RORC). However, the SAM/SAH ratios were somehow either positively or negatively correlated with the expressions of cytokines IL-17d (p<0.7196) and IL-17b (p<0.2739), respectively. These results suggest that higher cellular hypomethylation status as judged by the lower methylation status (SAM/SAH ratios) is positively associated with higher expressions of IL-17 receptors and lower expression of IL-17d.
Figure 5.
A. S-Adenosylhomocysteine (SAH) and S-Adenosylmethionine (SAM) Ratio is Associated with the Methylation Status of Tissues. B. Correlation Between IL-17 Cytokine and Receptor and Traf3ip2 and SAM/SAH Ratio in Mouse Tissues. Higher Hypomethylation Status is Positively Associated with Higher Expression of Gene. A) S-Adenosylhomocysteine (SAH) and S-Adenosylmethionine (SAM) are intermediate metabolites of the Homocysteine-methionine metabolism cycle. SAH is a potent inhibitor of cellular methylation; accumulation of SAH in tissues prevents methylation of DNA and other molecules by SAM. Abnormal DNA methylation has been reported to contribute to the development of cardiovascular and metabolic diseases. High SAM/SAH ratio in tissues is associated with hypermethylation of DNA and gene repression and low SAM/SAH ratio is associated with hypomethylation that leads to gene overexpression. B) Correlation of IL-17 cytokines, IL-17 receptors, and traf3ip2 expression with SAM/SAH ratio in mouse tissues. Relative expression of genes examined was determined as described in Figure 2 and expressed by relative mRNA expression levels. Tissue relative expressions of IL-17 cytokines, IL-17 receptors, and Traf3ip2 mRNA were plotted against tissue SAM/SAH ratios shown in Table 5. Linear regression analyses were performed using data points from 7 mouse tissues. Higher hypomethylation status (lower SAM/SAH ratio) is positively associated with higher expression of IL-17 receptors.
Table 5.
SAH and SAM levels found in mouse tissues
| Metabolite Concentrations (nmol/g wet wt) | |||
|---|---|---|---|
| Tissue | SAM | SAH | SAM:SAH |
| Brain | 35.8 ± 4.0 | 0.8 ± 0.1 | 47.1 ± 9.6 |
| Heart | 58.5 ± 4.2 | 0.4 ± 0.3 | 142.7 ± 87.6 |
| Kidney | 107.4 ± 5.5 | 4.2 ± 0.8 | 25.6 ± 5.0 |
| Liver | 112.8 ± 12.4 | 25.5 ± 3.9 | 4.4 ± 0.8 |
| Lung | 47.7± 3.6 | 5.5 ± 0.9 | 8.6 ± 1.5 |
| Spleen | 65.2 ± 8.1 | 1.7 ± 0.4 | 38.4 ± 10.2 |
| Thymus | 41.3 ± 11.5 | 1.2 ± 0.3 | 34.7 ± 13.6 |
Concentrations of SAM and SAH in mouse tissues were previously examined by Ueland et al.
4.5. RNA binding proteins may regulate the mRNA stability and translation of IL-17 cytokines and receptors
The results from tristetraprolin (TTP) knock-out mice and other studies suggest that TTP targets pro-inflammatory cytokine tumor necrosis factor (TNF)-alpha for decay in the exosome complex (54) via specific binding to AU-rich element (42) in the 3′UTR of TNF-alpha (35, 64). Since both TNF-alpha and IL-17 are pro-inflammatory cytokines, we hypothesized that mRNA stability mechanisms may regulate the mRNA stability of IL-17 cytokines and receptors. Using web-based AU-rich element mRNA database (65), we analyzed all the mRNA 3′UTRs of IL-17 cytokines and IL-17 receptors in the most comprehensive UTR database UTRdb (http://utrdb.ba.itb.cnr.it/search) (Table 6). As shown in Table 6, human IL-17A was the only molecule that contains an AU-rich element in the 3′UTR. In addition, other binding motifs for RNA binding proteins were found in some 3′UTRs of IL-17 cytokines and receptors. For example, Mos-PRE (Musashi Binding Element), which regulates the temporal order of mRNA translation (66), was found in the 3′UTR of human IL-17A mRNA. The GY-BOX (GTCTTCC)(67) was found in the 3′UTRs of human IL-17RB, IL-17RD, IL-17RE, and mouse IL-17rd mRNAs, respectively. Bearded (BRD)-BOX (AGCTTTA)(68) was found in the 3′UTRs of mouse IL-17a and IL-17d. BRD boxes and GY box confer negative regulatory activity on heterologous transcripts in vivo. UNR-bs (Upstream of N-ras Binding Site)(68) was found in the 3′UTR of mouse IL-17f and K-BOX (cTGTGATa)(69) was found in mouse IL-17rd. These results suggest that various RNA binding proteins may participate in regulating mRNA stability of IL-17 cytokines and receptors (36).
Table 6.
3′-Untranslated Region in mRNAs of IL-17 cytokines and receptors in human and mouse contain signals for RNA protein binding
| Human Gene | Length | Signal | Position | Mouse Gene | Length | Signal | Position |
|---|---|---|---|---|---|---|---|
| IL-17A | 1346 | Mos-PRE, ARE1 | 703–726, 261-14, 383–446, 624–628 | IL-17a | 637 | BRD-BOX | 462–468 |
| IL-17B | 102 | IL-17b | 100 | ||||
| IL-17C | 402 | IL-17c | 712 | ||||
| IL-17D | 1144 | IL-17d | 526 | BRD-BOX | 118–124 | ||
| IL-17E | 523 | IL-17e | 475 | ||||
| IL-17F | 245 | IL-17f | 621 | UNR-bs | 244–257 | ||
| IL-17RA | 695 | IL-17ra | 548 | ||||
| IL-17RB | 492 | GY-BOX | 293–299 | IL-17rb | 456 | ||
| IL-17RC | 37 | IL-17rc | 19 | ||||
| IL-17RD | 6411 | GY-BOX | 1019–1025 | IL-17rd | 5900 | K-BOX, GY-BOX | 3131–3138, 4166–4172, 4526–4532 |
| IL-17RE | 595 | GY-BOX | 242–248 | IL-17re | 608 | ||
| TRAF3IP2 | 465 | traf3ip2 | 778 |
Mos-PRE: Musashi Binding Element; ARE: AU-Rich Element; GY-BOX: GTCTTCC; BRD-BOX: AGCTTTA; UNR-bs: Upstream of N-ras Binding Site; K-BOX: cTGTGATa;
Class II ARE with 3 clusters. The data were retrieved from UTRdb (http://utrdb.ba.itb.cnr.it/search) at the Institute for Biomedical Technologies, University of Bar except that marked with 2, which was from the NIH-NCBI-AceView database.
4.6. MicroRNAs may regulate the mRNA stability and translation of IL-17 cytokines and receptors independently or via interaction with RNA binding protein-mediated mechanism
MicroRNAs (miRNAs or miRs) is a newly characterized class of short (18–24 nucleotide long)(70), endogenous and non-coding RNAs, which contribute to the development of particular disease states through the regulation of diverse biological processes such as cell growth, differentiation, proliferation, and apoptosis (37). This regulation occurs through base-pairing predominately with messenger RNAs (mRNAs) at the 3′UTR (71, 72), and leads to target mRNA cleavage and degradation or inhibition of mRNA translation (73). Sequence analysis identified miR-16 as possessing complementary sequence to the canonical AUUUA and demonstrated a role for this microRNA in interaction with the AU-rich element (74). Since we found an AU-rich element in the 3′UTR of human IL-17A, we hypothesized that mRNAs of IL-17 cytokines and receptors contain the structures in their 3′UTR for microRNA binding and regulation (Table 6). To examine this hypothesis, we used the online microRNA target prediction software, TargetScan (http://www.targetscan.org/) developed in MIT. The rationale for the use of this prediction database is presented in the discussion. To ensure that the predicted microRNAs have the binding quality equivalent to that of the experimentally verified microRNAs, we hypothesized that experimentally verified microRNAs have certain shared binding features between microRNAs and targeted 3′UTRs of mRNAs that are reflected in the context value and context percentage. To test this hypothesis, the confidence intervals for context value (the mean ± 2 x SD = -0.25 ± 0.12) and that for context percentage (76.07 ± 19.07) were generated, respectively, from the 45 interaction between 27 experimentally verified human microRNAs and 36 different genes (not shown) within the Tarbase, an online database of experimentally verified microRNAs (http://diana.cslab.ece.ntua.gr/tarbase/)(46, 47). These human microRNAs were confirmed using luciferase reporter assays and had effectively targeted a single unique mRNA sequence.
Using the microRNA target prediction software TargetScan, 909 microRNA binding sites were found in the 3′UTRs of the mRNAs of IL-17 cytokines and receptors. Using the confidence intervals of context value and context percentage, 125 microRNAs out of total predicted 909 microRNAs (125/909=13.75%) were selected to target the mRNAs of IL-17 cytokines and IL-17 receptors except IL-17RC (Table 7 and 8) with the binding quality equivalent to that of the experimentally verified microRNA-mRNA binding sites. The results suggest that the statistical confidence intervals are highly selective for the qualified microRNA-mRNA bindings. Among 125 high quality microRNAs, 39 selected microRNAs were found to target human IL-17A. Eight human IL-17A-targeting microRNAs, including miR-129-3p, miR-30b, miR-30c, miR-548a-5p, miR-548i, miR-548n and miR-938, targeted the same/nearby sequence region, bases 234–253 of the 3′UTR of human IL-17A. Similarly, three out of eight microRNAs targeted to the same sequence region (bases 216–225) in the 3′UTR of human IL-17C, and six out of nine microRNAs targeted to the same sequence region (bases 167–175) in 3′UTR of human IL-17F. These results suggest that the same sequence region in the 3′UTR of the mRNAs of IL-17 cytokines and receptors can be targeted by several different microRNAs, and that these “hot spots” in the 3′UTRs of mRNAs may be important for microRNA-mediated post-transcriptional regulation of IL-17 cytokine and receptor expression. In addition, some microRNAs were found to target more than one mRNA. For example, miR-129 targeted human IL-17A, IL-17D, and IL-17RB; miR-383 targeted human IL-17A and IL-17RD; and miR-1248 targeted human IL-17A and IL-17RD. This finding correlated well with others’ report that some microRNAs have numerous mRNA targets (75, 76). Furthermore, our analysis on experimental reports showed that 10 microRNAs, that target IL-17 cytokines and receptors, also targeted the mRNAs involved in cardiovascular disease, inflammatory molecule mRNAs, and cancer-related mRNAs (Table 9). The results suggest that signaling pathways regulating the expression and translation of IL-17 cytokines and receptors may be related to pathogenic processes of cardiovascular disease, inflammation, and cancer.
Table 7.
MicroRNA binding sites are found in the 3′UTR of IL-17 cytokine and receptor mRNAs
| Molecule | Total Predicted miR binding Sites | Significant predicted miR binding sites |
|---|---|---|
| IL-17A | 125 | 42 |
| IL-17B | 11 | 2 |
| IL-17C | 36 | 8 |
| IL-17D | 73 | 13 |
| IL-17E | 10 | 1 |
| IL-17F | 33 | 9 |
| IL-17RA | 58 | 4 |
| IL-17RB | 41 | 19 |
| IL-17RC | 2 | 0 |
| IL-17RD | 476 | 23 |
| IL-17RE | 44 | 4 |
Table 8.
MicroRNAs may regulate the mRNA translatability and mRNA stability of IL-17 cytokines and receptors via 3′untranslated region-dependent mechanisms
| mRNA | MicroRNA | Position on 3′UTR | mRNA | MicroRNA | Position on 3′UTR |
|---|---|---|---|---|---|
| IL-17A | hiR-1267 | 1142–1148 | IL-17B | miR-1271 | 55–61 |
| miR-1248 | 198–204 | miR-96 | 55–61 | ||
| miR-1266 | 279–285 | ||||
| miR-127-5p | 140–146 | IL-17C | miR-1184 | 217–223 | |
| miR-1290 | 702–708 | miR-1275 | 90–96 | ||
| miR-129-3p | 247–253 | miR-1301 | 216–222 | ||
| miR-1299 | 1304–1310 | miR-369-3p | 374–380 | ||
| miR-1324 | 1195–1201 | miR-485-5p | 164–170 | ||
| miR-142-5p | 326–332 | miR-544 | 219–225 | ||
| miR-146a | 747–753, 872–878 | miR-650 | 86–92 | ||
| miR-146b-5p | 747–753, 872–878 | miR-939 | 38–44 | ||
| miR-147 | 1211–1217 | ||||
| miR-30b | 234–240 | IL-17D | miR-1204 | 976–982 | |
| miR-30c | 234–240 | miR-1229 | 865–871 | ||
| miR-324-3p | 1236–1242 | miR-129-3p | 938–944 | ||
| miR-383 | 1198–1204 | miR-194 | 493–499 | ||
| miR-423-5p | 56–62, 1130–1136 | miR-220b | 862–868 | ||
| miR-485-5p | 85–91 | miR-324-5p | 713–719 | ||
| miR-507 | 512–518 | miR-331-5p | 423–429 | ||
| miR-515-5p | 1118–1124 | miR-423-3p | 99–105 | ||
| miR-520f | 460–466 | miR-548n | 428–434 | ||
| miR-548a-5p | 237–243 | miR-549 | 899–905 | ||
| miR-548i | 237–243 | miR-579 | 478–484 | ||
| miR-548n | 238–244 | miR-606 | 1103–1109 | ||
| miR-552 | 397–403 | miR-625 | 820–826 | ||
| miR-557 | 512–518 | ||||
| miR-559 | 237–243 | IL-17E | miR-370 | 116–122 | |
| miR-578 | 1046–1052 | ||||
| miR-618 | 110–116 | IL-17F | miR-106a | 167–173 | |
| miR-626 | 82–88 | miR-1257 | 140–146 | ||
| miR-629 | 688–694 | miR-1324 | 70–76 | ||
| miR-635 | 1271–1277 | miR-142-5p | 168–174 | ||
| miR-643 | 1044–1050 | miR-17 | 167–173 | ||
| miR-655 | 617–623 | miR-20a | 167–173 | ||
| miR-664 | 314–320 | miR-20b | 167–173 | ||
| miR-671-5p | 1110–1116 | miR-340 | 169–175 | ||
| miR-886-5p | 48–54 | miR-555 | 45–51 | ||
| miR-888 | 835–841 | ||||
| miR-938 | 246–252 | IL-17RD | miR-1236 | 1975–1981 | |
| miR-1248 | 4595–4601 | ||||
| IL-17RA | miR-331-3p | 197–203 | miR-1270 | 180–186 | |
| miR-377 | 99–105 | miR-134 | 570–576 | ||
| miR-597 | 542–548 | miR-182 | 242–248 | ||
| miR-661 | 143–149 | miR-21 | 530–536 | ||
| miR-330-3p | 396–402 | ||||
| IL-17RB | miR-1225-5p | 320–326 | miR-338-3p | 3773–3779 | |
| miR-1274a | 50–56 | miR-383 | 2063–2069 | ||
| miR-129-5p | 431–437 | miR-412 | 3139–3145 | ||
| miR-155 | 160–166 | miR-490-5p | 460–466 | ||
| miR-221 | 156–162 | miR-515-3p | 410–416 | ||
| miR-222 | 156–162 | miR-519d | 412–418 | ||
| miR-376c | 345–351 | miR-519e | 410–416 | ||
| miR-380 | 424–430 | miR-532-3p | 279–285 | ||
| miR-382 | 190–196 | miR-583 | 1003–1009 | ||
| miR-522 | 248–254 | miR-587 | 2342–2348 | ||
| miR-548a-3p | 227–233 | miR-589 | 483–489 | ||
| miR-548d-3p | 444–450 | miR-590-5p | 530–536 | ||
| miR-548e | 227–233 | miR-643 | 100–106 | ||
| miR-548f | 227–233 | miR-758 | 1035–1041 | ||
| miR-548g | 226–232 | miR-769-5p | 93–99 | ||
| miR-590-3p | 131–137 | miR-943 | 569–575 | ||
| miR-664 | 358–364 | ||||
| miR-671-5p | 30–36 | IL-17RE | miR-384 | 468–474 | |
| miR-7 | 294–300 | miR-511 | 569–575 | ||
| miR-516b | 258–264 | ||||
| IL-17RC | Not Identified | miR-516b | 266–272 |
Table 9.
Some IL-17 cytokine- and receptor-targeting MicroRNAs also target cardiovascular disease molecules, inflammation molecules, and cancer-related molecules
| MicroRNA | Target IL-17 Cytokines and Receptors | Target CVD Molecules | Target Inflammation Molecules | Target Cancer-Related Molecules |
|---|---|---|---|---|
| MiR-146 | IL-17A | TRAF6/IRAK-1(87), IRF-5(88) | ||
| MiR-17 | IL-17F | p21, Jak1(89) | AML-1(90) | |
| MiR-20a | IL-17F | VEGF(91) | AML-1(92) | TGFBR2(90) |
| MiR-20b | IL-17F | VEGF(92) | ||
| MiR-155 | IL-17RB | AT1R(93) | MMP-3(94), PU.1(95), TAB2(96) | |
| MiR-221 | IL-17RB | c-Kit(97), eNOS(98) | ||
| MiR-222 | IL-17RB | c-Kit(97), eNOS(98) | ||
| MiR-21 | IL-17RD | RECK, TIMP3(99), TPM1(100), PTEN(101), PDCD4(102) |
5. DISCUSSION
IL-17 cytokines are a family of pro-inflammatory autoimmune cytokines (also see our invited review (12)). Despite significant progress, several important knowledge gaps exist which prevent investigators from defining the detailed roles of these molecules in inflammation and immune responses. Our current studies have made the following findings: i) most IL-17 cytokines are not constitutively expressed in the 16 tissues examined, but several IL-17 receptors and TRAF3IP2 are ubiquitously expressed with a few exceptions, suggesting the upregulation of IL-17 cytokines in response to pro-inflammatory stimuli is one of the major mechanisms for IL-17 signaling; ii) heart and vascular tissue are in the second tier of readiness to respond to IL-17 cytokine stimulation, which require the upregulation of IL-17 receptor components; iii) alternative promoters and alternative spliced isoforms are found in the transcripts of IL-17 cytokines and receptors, suggesting that tissue-specific and stimulation-specific alternative promoters and alternative splicing regulate the structures and expressions of IL-17 cytokines and receptors; iv) Higher hypomethylation status as judged by the lower SAM/SAH ratios is positively associated with higher expressions of several IL-17 receptors and lower expression of IL-17d in mouse tissues, suggesting that the expression of these molecules is also regulated by epigenetic methylation mechanism; v) An AU-rich element is found in the 3′UTR of human IL-17A and the binding sites of several RNA binding proteins are found in the 3′UTR of IL-17 cytokines and receptors, suggesting that RNA binding proteins may regulate the mRNA stability and translation of IL-17 cytokines and receptors; and vi) using the statistical confidence intervals generated with experimentally verified microRNA-mRNA binding sites, 125 binding sites in the mRNAs of IL-17 cytokines and receptors for qualified microRNAs to target were selected out of the 909 total predicted binding sites. The finding of microRNA binding sites in the 3′UTR of IL-17 cytokines and receptors statistically equivalent to that of experimentally verified microRNAs-mRNA interaction suggests that microRNAs may regulate the mRNA stability and translation of IL-17 cytokines and receptors. Of note, the microRNA-mediated regulations of mRNA stability of IL-17 cytokines and receptors are realized independently or via interaction with RNA binding protein-mediated mechanism.
It is worth to point out that the expression data retrieved from the expression sequence tag (EST) database analyzed in this study are more precise than that detected with traditional approaches including Northern blot analysis and PCR analysis due to the un-biased cDNA cloning and DNA sequencing procedures of EST database deposits (77). Thus, the expression patterns of IL-17 cytokines and receptors are experimentally based and precise.
Alternative promoters play an important role for gene transcription in response to tissue/cell-specific, and/or stimulation-specific transcription signaling (78). One of the best examples about multiple promoter usage is fibroblast growth factor-1 (FGF1) transcription, which is controlled by at least four distinct promoters in a tissue-specific manner. The 1.A and 1.B promoters of FGF1 are constitutively active in their respective cell types. In contrast, different biological response modifiers, including serum and transforming growth factor-beta, can induce the 1.C and 1.D promoters of FGF1(78). Identification of alternative promoters in human IL-17B, IL-17D, and IL-17F genes suggests that the transcription of these three human IL-17 cytokines may be under regulation of tissue-specific and/or stimulation-specific transcription signaling.
Post-transcriptional regulation controls the abundance, turnover, and translation of mRNA and offers the capacity to integrate signal transduction events with very rapid changes in gene expression during cellular differentiation, which can be realized via different mechanisms including RNA binding proteins such as Tristetraprolin family proteins and microRNAs (79). Previous report showed that mitogen-activated protein kinase (MAPK) stabilizes mRNAs through the inhibition of stabilizing proteins such as tristetraprolin. Tristetraprolin binds to AU-rich elements in mRNA transcripts and delivers them to the exosome complex, where they are degraded (54). Identification of AU-rich element in the 3′UTR of human IL-17A mRNA suggest that the mRNA stability of human IL-17A may be under regulation of tristetraprolin-activated MAPK pathway.
RNA binding proteins may also interact with microRNAs through mechanisms that are not fully understood, and there is evidence that both mechanisms can target the same mRNA(80). MicroRNAs are a newly characterized class of short (18–24 nucleotide long)(70), endogenous, and non-coding RNAs, which are processed by nuclear RNase Drosha and cytosolic RNase Dicer. MicroRNAs are capable of controlling complex biological functions through post-transcriptional gene silencing (73). A recent report showed that miRNA-326 promotes Th-17 cell differentiation by targeting Ets-1, a negative regulator of Th-17 cell differentiation (81), suggesting a possibility that IL-17 cytokines and receptors are under regulation of microRNAs. However, the issue of whether microRNAs regulate the expression of IL-17 cytokines and receptors remains unknown. Using the most widely used target prediction program TargetScan (http://www.targetscan.org/) (82–84), we predicted a list of microRNAs that could target the 3′UTRs of IL-17 cytokines and receptors mRNAs. We hypothesized that experimentally verified microRNAs have certain shared binding features between the microRNAs and the targeted 3′UTRs of mRNAs that are reflected in the context value and context percentage, which can further be analyzed with statistical methods. By examining this hypothesis, the confidence intervals were generated, which allowed us to identify the microRNAs with the binding features statistically equivalent to that of experimentally verified microRNA-mRNA interaction. In our study, introduction of statistical method to generate the confidence intervals of experimentally verified microRNA-mRNA binding features sort out a small portion of high quality microRNAs among total predicted ones and significantly improves our prediction. Our result suggest that microRNAs as a new mechanism may regulate the mRNA stability and translation of IL-17 cytokines and receptors. This software was used in this study based on the following rationales: first, this software includes conserved and poorly conserved miRNAs; second, it individually ranks microRNA-target mRNA binding efficacy; and third, this software is the most widely used target prediction program (82–84). Of note, some microRNAs that we predicted for targeting the mRNAs of IL-17 cytokines and receptors are well-characterized. Our data showed that miRNA-21 is found to target human IL-17RD. Previous reports showed that miRNA-21 has been validated as a bona fide oncogene, which decreases apoptosis, promotes survival and proliferation. In addition, one of the potential targets of miRNA-21 is IL-12p35, a subunit of IL-12, suggesting that miRNA-21 may regulate inflammation and type 1 T helper cell (Th1) polarization (85). Previous report also showed that miRNA-221/222 inhibits angiogenesis (86). Our results showed that miRNA-221/222 may inhibit human IL-17RB expression, implying that IL-17RB promotes angiogenesis. Moreover, previous report showed that miRNA-146 regulates several inflammatory pathways as part of a negative feedback (85). Our results suggest that miRNA-146 may inhibit human IL-17A expression, which correlated well with the previous studies. Finally, previous report showed that miRNA-155 promotes regulatory T cell development and suppresses excessive inflammation (85). Our results suggest that miRNA-155 may inhibit human IL-17RB expression, which correlated well with the previous studies. Taken together, our results correlated well with the previous reports for some well-characterized microRNAs with the function in regulating inflammation. In addition, eight human microRNAs, that target IL-17 cytokines and receptors, also targeted cardiovascular disease molecule mRNAs, inflammatory molecule mRNAs, and cancer-related mRNAs. In conclusion, as shown in our working model (Figure 6), our results suggest that microRNAs and other mechanisms regulate the expression, structure, and translation of IL-17 cytokines and receptors. These findings provide an insight into pathogenic processes of cardiovascular disease, inflammation, and cancers.
Figure 6.
Working Model of IL-17 Cytokine and Receptor Expression and Structure Regulations. Alternative promoters, alternative splicing, and post-transcriptional modifications all contribute to IL-17 cytokine and receptor expression and structure regulations. Approximately 50% of IL-17 cytokine and receptor genes contain multiple promoters which may have important role in tissue-specific and/or stimulation specific transcriptional regulation. Alternative splicing affects the open reading frames (ORF) of RNA transcript which are translated into different protein isoforms. RNA transcripts are regulated by post-transcripitonal modifications including RNA binding protein and microRNA interactions. Interactions with RNA binding protein and microRNA can enhance RNA stability or induce RNA destabilization leading to protein translation or RNA degradation, respectively.
Acknowledgments
Anthony Virtue, Erin Maley, and Tran Tran made equal contributions to this work. This work was partially supported by the National Institutes of Health Grants HL094451 (XFY), HL67033, HL82774, and HL77288 (HW).
References
- 1.Packard RR, Lichtman AH, Libby P. Innate and adaptive immunity in atherosclerosis. Semin Immunopathol. 2009;31:5–22. doi: 10.1007/s00281-009-0153-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ait-Oufella H, Salomon BL, Potteaux S, Robertson AK, Gourdy P, Zoll J, Merval R, Esposito B, Cohen JL, Fisson S, Flavell RA, Hansson GK, Klatzmann D, Tedgui A, Mallat Z. Natural regulatory T cells control the development of atherosclerosis in mice. Nat Med. 2006;12:178–80. doi: 10.1038/nm1343. [DOI] [PubMed] [Google Scholar]
- 3.Xiong Z, Song J, Yan Y, Huang Y, Cowan A, Wang H, Yang XF. Higher expression of Bax in regulatory T cells increases vascular inflammation. Front Biosci. 2008;13:7143–55. doi: 10.2741/3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiong Z, Yan Y, Song J, Fang P, Yin Y, Yang Y, Cowan A, Wang H, Yang XF. Expression of TCTP antisense in CD25 (high) regulatory T cells aggravates cuff-injured vascular inflammation. Atherosclerosis. 2009;203:401–8. doi: 10.1016/j.atherosclerosis.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang D, Jiang X, Fang P, Yan Y, Song J, Gupta S, Schafer AI, Durante W, Kruger WD, Yang X, Wang H. Hyperhomocysteinemia promotes inflammatory monocyte generation and accelerates atherosclerosis in transgenic cystathionine beta-synthase-deficient mice. Circulation. 2009;120:1893–902. doi: 10.1161/CIRCULATIONAHA.109.866889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fouser LA, Wright JF, Dunussi-Joannopoulos K, Collins M. Th17 cytokines and their emerging roles in inflammation and autoimmunity. Immunol Rev. 2008;226:87–102. doi: 10.1111/j.1600-065X.2008.00712.x. [DOI] [PubMed] [Google Scholar]
- 7.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 8.van Es T, van Puijvelde GH, Ramos OH, Segers FM, Joosten LA, van den Berg WB, Michon IM, de Vos P, van Berkel TJ, Kuiper J. Attenuated atherosclerosis upon IL-17R signaling disruption in LDLr deficient mice. Biochem Biophys Res Commun. 2009;388:261–5. doi: 10.1016/j.bbrc.2009.07.152. [DOI] [PubMed] [Google Scholar]
- 9.Xie JJ, Wang J, Tang TT, Chen J, Gao XL, Yuan J, Zhou ZH, Liao MY, Yao R, Yu X, Wang D, Cheng Y, Liao YH, Cheng X. The Th17/Treg functional imbalance during atherogenesis in ApoE (−/−) mice Th17/Treg imbalance in atherogenesis. Cytokine. 2009 doi: 10.1016/j.cyto.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 10.Taleb S, Romain M, Ramkhelawon B, Uyttenhove C, Pasterkamp G, Herbin O, Esposito B, Perez N, Yasukawa H, Van Snick J, Yoshimura A, Tedgui A, Mallat Z. Loss of SOCS3 expression in T cells reveals a regulatory role for interleukin-17 in atherosclerosis. J Exp Med. 2009;206:2067–77. doi: 10.1084/jem.20090545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erbel C, Klingenberg R, Celik S, Wambsganss N, Funke B, Boeckler D, Dengler T. Abstract 759: Inhibition of proinflammatory cytokine IL-17 reduces atherosclerotic lesion development in ApoE−/− mice. Circulation. 2007;116:145–146. [Google Scholar]
- 12.Mai J, Wang H, Yang X-FT. Helper 17 Cells Interplay with CD4+CD25highFoxp3+ Tregs in Regulation of Inflammations and Autoimmune Diseases. Frontiers in Bioscience. 2010 doi: 10.2741/3657. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Syrjala SO, Keranen MA, Tuuminen R, Nykanen AI, Tammi M, Krebs R, Lemstrom KB. Increased Th17 rather than Th1 alloimmune response is associated with cardiac allograft vasculopathy after hypothermic preservation in the rat. J Heart Lung Transplant. 2010 doi: 10.1016/j.healun.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 14.Kolls JK, Linden A. Interleukin-17 family members and inflammation. Immunity. 2004;21:467–76. doi: 10.1016/j.immuni.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 15.Rouvier E, Luciani MF, Mattei MG, Denizot F, Golstein P. CTLA-8, cloned from an activated T cell, bearing AU-rich messenger RNA instability sequences, and homologous to a herpesvirus saimiri gene. J Immunol. 1993;150:5445–56. [PubMed] [Google Scholar]
- 16.Yao Z, Painter SL, Fanslow WC, Ulrich D, Macduff BM, Spriggs MK, Armitage RJ. Human IL-17: a novel cytokine derived from T cells. J Immunol. 1995;155:5483–6. [PubMed] [Google Scholar]
- 17.Li H, Chen J, Huang A, Stinson J, Heldens S, Foster J, Dowd P, Gurney AL, Wood WI. Cloning and characterization of IL-17B and IL-17C, two new members of the IL-17 cytokine family. Proc Natl Acad Sci U S A. 2000;97:773–8. doi: 10.1073/pnas.97.2.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J, Ho WH, Maruoka M, Corpuz RT, Baldwin DT, Foster JS, Goddard AD, Yansura DG, Vandlen RL, Wood WI, Gurney AL. IL-17E, a novel proinflammatory ligand for the IL-17 receptor homolog IL-17Rh1. J Biol Chem. 2001;276:1660–4. doi: 10.1074/jbc.M008289200. [DOI] [PubMed] [Google Scholar]
- 19.Hymowitz SG, Filvaroff EH, Yin JP, Lee J, Cai L, Risser P, Maruoka M, Mao W, Foster J, Kelley RF, Pan G, Gurney AL, de Vos AM, Starovasnik MA. IL-17s adopt a cystine knot fold: structure and activity of a novel cytokine, IL-17F, and implications for receptor binding. Embo J. 2001;20:5332–41. doi: 10.1093/emboj/20.19.5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Starnes T, Robertson MJ, Sledge G, Kelich S, Nakshatri H, Broxmeyer HE, Hromas R. Cutting edge: IL-17F, a novel cytokine selectively expressed in activated T cells and monocytes, regulates angiogenesis and endothelial cell cytokine production. J Immunol. 2001;167:4137–40. doi: 10.4049/jimmunol.167.8.4137. [DOI] [PubMed] [Google Scholar]
- 21.Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, Wang Y, Hood L, Zhu Z, Tian Q, Dong C. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6:1133–41. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawaguchi M, Kokubu F, Odaka M, Watanabe S, Suzuki S, Ieki K, Matsukura S, Kurokawa M, Adachi M, Huang SK. Induction of granulocyte-macrophage colony-stimulating factor by a new cytokine, ML-1 (IL-17F), via Raf I-MEK-ERK pathway. J Allergy Clin Immunol. 2004;114:444–50. doi: 10.1016/j.jaci.2004.03.047. [DOI] [PubMed] [Google Scholar]
- 23.Awane M, Andres PG, Li DJ, Reinecker HC. NF-kappa B-inducing kinase is a common mediator of IL-17-, TNF-alpha-, and IL-1 beta-induced chemokine promoter activation in intestinal epithelial cells. J Immunol. 1999;162:5337–44. [PubMed] [Google Scholar]
- 24.Shalom-Barak T, Quach J, Lotz M. Interleukin-17-induced gene expression in articular chondrocytes is associated with activation of mitogen-activated protein kinases and NF-kappaB. J Biol Chem. 1998;273:27467–73. doi: 10.1074/jbc.273.42.27467. [DOI] [PubMed] [Google Scholar]
- 25.Chang SH, Dong C. A novel heterodimeric cytokine consisting of IL-17 and IL-17F regulates inflammatory responses. Cell Res. 2007;17:435–40. doi: 10.1038/cr.2007.35. [DOI] [PubMed] [Google Scholar]
- 26.Moseley TA, Haudenschild DR, Rose L, Reddi AH. Interleukin-17 family and IL-17 receptors. Cytokine Growth Factor Rev. 2003;14:155–74. doi: 10.1016/s1359-6101(03)00002-9. [DOI] [PubMed] [Google Scholar]
- 27.Yao Z, Fanslow WC, Seldin MF, Rousseau AM, Painter SL, Comeau MR, Cohen JI, Spriggs MK. Herpesvirus Saimiri encodes a new cytokine, IL-17, which binds to a novel cytokine receptor. Immunity. 1995;3:811–21. doi: 10.1016/1074-7613(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 28.Kuestner RE, Taft DW, Haran A, Brandt CS, Brender T, Lum K, Harder B, Okada S, Ostrander CD, Kreindler JL, Aujla SJ, Reardon B, Moore M, Shea P, Schreckhise R, Bukowski TR, Presnell S, Guerra-Lewis P, Parrish-Novak J, Ellsworth JL, Jaspers S, Lewis KE, Appleby M, Kolls JK, Rixon M, West JW, Gao Z, Levin SD. Identification of the IL-17 receptor related molecule IL-17RC as the receptor for IL-17F. J Immunol. 2007;179:5462–73. doi: 10.4049/jimmunol.179.8.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toy D, Kugler D, Wolfson M, Vanden Bos T, Gurgel J, Derry J, Tocker J, Peschon J. Cutting edge: interleukin 17 signals through a heteromeric receptor complex. J Immunol. 2006;177:36–9. doi: 10.4049/jimmunol.177.1.36. [DOI] [PubMed] [Google Scholar]
- 30.Gratchev A, Kzhyshkowska J, Duperrier K, Utikal J, Velten FW, Goerdt S. The receptor for interleukin-17E is induced by Th2 cytokines in antigen-presenting cells. Scand J Immunol. 2004;60:233–7. doi: 10.1111/j.0300-9475.2004.01443.x. [DOI] [PubMed] [Google Scholar]
- 31.Novatchkova M, Leibbrandt A, Werzowa J, Neubuser A, Eisenhaber F. The STIR-domain superfamily in signal transduction, development and immunity. Trends Biochem Sci. 2003;28:226–9. doi: 10.1016/S0968-0004(03)00067-7. [DOI] [PubMed] [Google Scholar]
- 32.Dong C. Regulation and pro-inflammatory function of interleukin-17 family cytokines. Immunol Rev. 2008;226:80–6. doi: 10.1111/j.1600-065X.2008.00709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Linden A. A role for the cytoplasmic adaptor protein Act1 in mediating IL-17 signaling. Sci STKE. 2007;2007:re4. doi: 10.1126/stke.3982007re4. [DOI] [PubMed] [Google Scholar]
- 34.Qian Y, Liu C, Hartupee J, Altuntas CZ, Gulen MF, Jane-Wit D, Xiao J, Lu Y, Giltiay N, Liu J, Kordula T, Zhang QW, Vallance B, Swaidani S, Aronica M, Tuohy VK, Hamilton T, Li X. The adaptor Act1 is required for interleukin 17-dependent signaling associated with autoimmune and inflammatory disease. Nat Immunol. 2007;8:247–56. doi: 10.1038/ni1439. [DOI] [PubMed] [Google Scholar]
- 35.Sandler H, Stoecklin G. Control of mRNA decay by phosphorylation of tristetraprolin. Biochem Soc Trans. 2008;36:491–6. doi: 10.1042/BST0360491. [DOI] [PubMed] [Google Scholar]
- 36.Anderson P. Post-transcriptional control of cytokine production. Nat Immunol. 2008;9:353–9. doi: 10.1038/ni1584. [DOI] [PubMed] [Google Scholar]
- 37.Zhang C. MicroRNAs: role in cardiovascular biology and disease. Clin Sci (Lond) 2008;114:699–706. doi: 10.1042/CS20070211. [DOI] [PubMed] [Google Scholar]
- 38.Ng B, Yang F, Huston DP, Yan Y, Yang Y, Xiong Z, Peterson LE, Wang H, Yang XF. Increased noncanonical splicing of autoantigen transcripts provides the structural basis for expression of untolerized epitopes. J Allergy Clin Immunol. 2004;114:1463–70. doi: 10.1016/j.jaci.2004.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen NC, Yang F, Capecci LM, Gu Z, Schafer AI, Durante W, Yang XF, Wang H. Regulation of homocysteine metabolism and methylation in human and mouse tissues. Faseb J. 2010;24:2804–17. doi: 10.1096/fj.09-143651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin Y, Yan Y, Jiang X, Mai J, Chen NC, Wang H, Yang XF. Inflammasomes are differentially expressed in cardiovascular and other tissues. Int J Immunopathol Pharmacol. 2009;22:311–22. doi: 10.1177/039463200902200208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pires Martins R, Leach RE, Krawetz SA. Whole-body gene expression by data mining. Genomics. 2001;72:34–42. doi: 10.1006/geno.2000.6437. [DOI] [PubMed] [Google Scholar]
- 42.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–70. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 43.Ueland PM, Helland S, Broch OJ, Schanche JS. Homocysteine in tissues of the mouse and rat. J Biol Chem. 1984;259:2360–4. [PubMed] [Google Scholar]
- 44.Helland S, Ueland PM. S-adenosylhomocysteine and S-adenosylhomocysteine hydrolase in various tissues of mice given injections of 9-beta-D-arabinofuranosyladenine. Cancer Res. 1983;43:1847–50. [PubMed] [Google Scholar]
- 45.Helland S, Ueland PM. Effect of 2′-deoxycoformycin infusion on S-adenosylhomocysteine hydrolase and the amount of S-adenosylhomocysteine and related compounds in tissues of mice. Cancer Res. 1983;43:4142–7. [PubMed] [Google Scholar]
- 46.Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: A comprehensive database of experimentally supported animal microRNA targets. Rna. 2006;12:192–7. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Papadopoulos GL, Reczko M, Simossis VA, Sethupathy P, Hatzigeorgiou AG. The database of experimentally supported targets: a functional update of TarBase. Nucleic Acids Res. 2009;37:D155–8. doi: 10.1093/nar/gkn809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Starnes T, Broxmeyer HE, Robertson MJ, Hromas R. Cutting edge: IL-17D, a novel member of the IL-17 family, stimulates cytokine production and inhibits hemopoiesis. J Immunol. 2002;169:642–6. doi: 10.4049/jimmunol.169.2.642. [DOI] [PubMed] [Google Scholar]
- 49.Silva WA, Jr, Covas DT, Panepucci RA, Proto-Siqueira R, Siufi JL, Zanette DL, Santos AR, Zago MA. The profile of gene expression of human marrow mesenchymal stem cells. Stem Cells. 2003;21:661–9. doi: 10.1634/stemcells.21-6-661. [DOI] [PubMed] [Google Scholar]
- 50.Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, Weaver CT. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6:1123–32. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 51.Weaver CT, Harrington LE, Mangan PR, Gavrieli M, Murphy KM. Th17: an effector CD4 T cell lineage with regulatory T cell ties. Immunity. 2006;24:677–88. doi: 10.1016/j.immuni.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 52.Claudio E, Sonder SU, Saret S, Carvalho G, Ramalingam TR, Wynn TA, Chariot A, Garcia-Perganeda A, Leonardi A, Paun A, Chen A, Ren NY, Wang H, Siebenlist U. The adaptor protein CIKS/Act1 is essential for IL-25-mediated allergic airway inflammation. J Immunol. 2009;182:1617–30. doi: 10.4049/jimmunol.182.3.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang SH, Park H, Dong C. Act1 adaptor protein is an immediate and essential signaling component of interleukin-17 receptor. J Biol Chem. 2006;281:35603–7. doi: 10.1074/jbc.C600256200. [DOI] [PubMed] [Google Scholar]
- 54.Gaffen SL. Structure and signalling in the IL-17 receptor family. Nat Rev Immunol. 2009;9:556–67. doi: 10.1038/nri2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Letuve S, Lajoie-Kadoch S, Audusseau S, Rothenberg ME, Fiset PO, Ludwig MS, Hamid Q. IL-17E upregulates the expression of proinflammatory cytokines in lung fibroblasts. J Allergy Clin Immunol. 2006;117:590–6. doi: 10.1016/j.jaci.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 56.Kornblihtt AR. Promoter usage and alternative splicing. Curr Opin Cell Biol. 2005;17:262–8. doi: 10.1016/j.ceb.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 57.Kornblihtt AR. Coupling transcription and alternative splicing. Adv Exp Med Biol. 2007;623:175–89. doi: 10.1007/978-0-387-77374-2_11. [DOI] [PubMed] [Google Scholar]
- 58.Thierry-Mieg D, Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7(Suppl 1):S12 1–14. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Akimzhanov AM, Yang XO, Dong C. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J Biol Chem. 2007;282:5969–72. doi: 10.1074/jbc.C600322200. [DOI] [PubMed] [Google Scholar]
- 60.Ordovas JM, Smith CE. Epigenetics and cardiovascular disease. Nat Rev Cardiol. 2010 doi: 10.1038/nrcardio.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jamaluddin MS, Yang X, Wang H. Hyperhomocysteinemia, DNA methylation and vascular disease. Clin Chem Lab Med. 2007;45:1660–6. doi: 10.1515/CCLM.2007.350. [DOI] [PubMed] [Google Scholar]
- 62.James SJ, Melnyk S, Pogribna M, Pogribny IP, Caudill MA. Elevation in S-adenosylhomocysteine and DNA hypomethylation: potential epigenetic mechanism for homocysteine-related pathology. J Nutr. 2002;132:2361S–2366S. doi: 10.1093/jn/132.8.2361S. [DOI] [PubMed] [Google Scholar]
- 63.Caudill MA, Wang JC, Melnyk S, Pogribny IP, Jernigan S, Collins MD, Santos-Guzman J, Swendseid ME, Cogger EA, James SJ. Intracellular S-adenosylhomocysteine concentrations predict global DNA hypomethylation in tissues of methyl-deficient cystathionine beta-synthase heterozygous mice. J Nutr. 2001;131:2811–8. doi: 10.1093/jn/131.11.2811. [DOI] [PubMed] [Google Scholar]
- 64.Taylor GA, Carballo E, Lee DM, Lai WS, Thompson MJ, Patel DD, Schenkman DI, Gilkeson GS, Broxmeyer HE, Haynes BF, Blackshear PJ. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4:445–54. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- 65.Bakheet T, Williams BR, Khabar KS. ARED 2.0: an update of AU-rich element mRNA database. Nucleic Acids Res. 2003;31:421–3. doi: 10.1093/nar/gkg023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Charlesworth A, Wilczynska A, Thampi P, Cox LL, MacNicol AM. Musashi regulates the temporal order of mRNA translation during Xenopus oocyte maturation. Embo J. 2006;25:2792–801. doi: 10.1038/sj.emboj.7601159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lai EC, Posakony JW. The Bearded box, a novel 3′ UTR sequence motif, mediates negative post-transcriptional regulation of Bearded and Enhancer of split Complex gene expression. Development. 1997;124:4847–56. doi: 10.1242/dev.124.23.4847. [DOI] [PubMed] [Google Scholar]
- 68.Dinur M, Kilav R, Sela-Brown A, Jacquemin-Sablon H, Naveh-Many T. In vitro evidence that upstream of N-ras participates in the regulation of parathyroid hormone messenger ribonucleic acid stability. Mol Endocrinol. 2006;20:1652–60. doi: 10.1210/me.2005-0333. [DOI] [PubMed] [Google Scholar]
- 69.Lai EC, Burks C, Posakony JW. The K box, a conserved 3′ UTR sequence motif, negatively regulates accumulation of enhancer of split complex transcripts. Development. 1998;125:4077–88. doi: 10.1242/dev.125.20.4077. [DOI] [PubMed] [Google Scholar]
- 70.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rasmussen KD, Simmini S, Abreu-Goodger C, Bartonicek N, Di Giacomo M, Bilbao-Cortes D, Horos R, Von Lindern M, Enright AJ, O’Carroll D. The miR-144/451 locus is required for erythroid homeostasis. J Exp Med. 2010;207:1351–8. doi: 10.1084/jem.20100458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–10. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Naeem H, Kuffner R, Csaba G, Zimmer R. miRSel: automated extraction of associations between microRNAs and genes from the biomedical literature. BMC Bioinformatics. 2010;11:135. doi: 10.1186/1471-2105-11-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di Padova F, Lin SC, Gram H, Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–34. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 75.Asirvatham AJ, Gregorie CJ, Hu Z, Magner WJ, Tomasi TB. MicroRNA targets in immune genes and the Dicer/Argonaute and ARE machinery components. Mol Immunol. 2008;45:1995–2006. doi: 10.1016/j.molimm.2007.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Asirvatham AJ, Magner WJ, Tomasi TB. miRNA regulation of cytokine genes. Cytokine. 2009;45:58–69. doi: 10.1016/j.cyto.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yin Y, Yan Y, Jiang X, Mai J, Chen NC, Wang H, Yang XF. Inflammasomes are differentially expressed in cardiovascular and other tissues. Int J Immunopathol Pharmacol. 2009;22:311–322. doi: 10.1177/039463200902200208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chiu IM, Touhalisky K, Baran C. Multiple controlling mechanisms of FGF1 gene expression through multiple tissue-specific promoters. Prog Nucleic Acid Res Mol Biol. 2001;70:155–74. doi: 10.1016/s0079-6603(01)70016-5. [DOI] [PubMed] [Google Scholar]
- 79.Ademokun A, Turner M. Regulation of B-cell differentiation by microRNAs and RNA-binding proteins. Biochem Soc Trans. 2008;36:1191–3. doi: 10.1042/BST0361191. [DOI] [PubMed] [Google Scholar]
- 80.Kedde M, Agami R. Interplay between microRNAs and RNA-binding proteins determines developmental processes. Cell Cycle. 2008;7:899–903. doi: 10.4161/cc.7.7.5644. [DOI] [PubMed] [Google Scholar]
- 81.Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, Li Z, Wu Z, Pei G. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009;10:1252–9. doi: 10.1038/ni.1798. [DOI] [PubMed] [Google Scholar]
- 82.Vickers KC, Remaley AT. MicroRNAs in atherosclerosis and lipoprotein metabolism. Curr Opin Endocrinol Diabetes Obes. 2010;17:150–5. doi: 10.1097/MED.0b013e32833727a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rosero S, Bravo-Egana V, Jiang Z, Khuri S, Tsinoremas N, Klein D, Sabates E, Correa-Medina M, Ricordi C, Dominguez-Bendala J, Diez J, Pastori RL. MicroRNA signature of the human developing pancreas. BMC Genomics. 2010;11:509. doi: 10.1186/1471-2164-11-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dong H, Paquette M, Williams A, Zoeller RT, Wade M, Yauk C. Thyroid hormone may regulate mRNA abundance in liver by acting on microRNAs. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sonkoly E, Pivarcsi A. microRNAs in inflammation. Int Rev Immunol. 2009;28:535–61. doi: 10.3109/08830180903208303. [DOI] [PubMed] [Google Scholar]
- 86.Haver VG, Slart RH, Zeebregts CJ, Peppelenbosch MP, Tio RA. Rupture of vulnerable atherosclerotic plaques: microRNAs conducting the orchestra? Trends Cardiovasc Med. 2010;20:65–71. doi: 10.1016/j.tcm.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 87.Nakasa T, Miyaki S, Okubo A, Hashimoto M, Nishida K, Ochi M, Asahara H. Expression of microRNA-146 in rheumatoid arthritis synovial tissue. Arthritis Rheum. 2008;58:1284–92. doi: 10.1002/art.23429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tang Y, Luo X, Cui H, Ni X, Yuan M, Guo Y, Huang X, Zhou H, de Vries N, Tak PP, Chen S, Shen N. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009;60:1065–75. doi: 10.1002/art.24436. [DOI] [PubMed] [Google Scholar]
- 89.Doebele C, Bonauer A, Fischer A, Scholz A, Reiss Y, Urbich C, Hofmann WK, Zeiher AM, Dimmeler S. Members of the microRNA-17–92 cluster exhibit a cell-intrinsic antiangiogenic function in endothelial cells. Blood. 2010;115:4944–50. doi: 10.1182/blood-2010-01-264812. [DOI] [PubMed] [Google Scholar]
- 90.Fontana L, Pelosi E, Greco P, Racanicchi S, Testa U, Liuzzi F, Croce CM, Brunetti E, Grignani F, Peschle C. MicroRNAs 17–5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat Cell Biol. 2007;9:775–87. doi: 10.1038/ncb1613. [DOI] [PubMed] [Google Scholar]
- 91.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hua Z, Lv Q, Ye W, Wong CK, Cai G, Gu D, Ji Y, Zhao C, Wang J, Yang BB, Zhang Y. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One. 2006;1:e116. doi: 10.1371/journal.pone.0000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pauley KM, Satoh M, Chan AL, Bubb MR, Reeves WH, Chan EK. Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Res Ther. 2008;10:R101. doi: 10.1186/ar2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Martinez-Nunez RT, Louafi F, Friedmann PS, Sanchez-Elsner T. MicroRNA-155 modulates the pathogen binding ability of dendritic cells (DCs) by down-regulation of DC-specific intercellular adhesion molecule-3 grabbing non-integrin (DC-SIGN) J Biol Chem. 2009;284:16334–42. doi: 10.1074/jbc.M109.011601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ceppi M, Pereira PM, Dunand-Sauthier I, Barras E, Reith W, Santos MA, Pierre P. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc Natl Acad Sci U S A. 2009;106:2735–40. doi: 10.1073/pnas.0811073106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Martin MM, Lee EJ, Buckenberger JA, Schmittgen TD, Elton TS. MicroRNA-155 regulates human angiotensin II type 1 receptor expression in fibroblasts. J Biol Chem. 2006;281:18277–84. doi: 10.1074/jbc.M601496200. [DOI] [PubMed] [Google Scholar]
- 97.Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, Woods K, Mercatanti A, Hammond S, Rainaldi G. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108:3068–71. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 98.Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res. 2007;100:1164–73. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- 99.Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, Krichevsky AM. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28:5369–80. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282:14328–36. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 101.Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, Jiang J, Schmittgen TD, Patel T. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–29. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 102.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–36. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]