Abstract
The structure and connectivity of protein-protein interaction (PPI) networks are maintained throughout evolution by coordinated changes (coevolution) of network proteins. Despite extensive research, relatively little is known regarding the molecular basis and functional implications of the coevolution of PPI networks. Here, we used proliferating cell nuclear antigen, a hub protein that mediates DNA replication and repair in eukaryotes, as a model system to study the coevolution of PPI networks in fungi. Using a combined bioinformatics and experimental approach, we discovered that PCNA-partner interactions tightly coevolved in fungal species, leading to specific modes of recognition. We found that fungal proliferating cell nuclear antigen-partner interaction networks diverged into two distinct groups as a result of such coevolution and that hybrid networks of these groups are functionally noncompatible in Saccharomyces cerevisiae. Our results indicate that the coevolution of PPI networks can form functional barriers between fungal species, and thus can promote and fix speciation.
Keywords: interdomain connecting loop, yeast two hybrid, directed evolution
Protein-protein interaction (PPI) networks play vital roles in executing almost all essential biological processes. The availability of sequencing data, as well as high-throughput experimental approaches to identify and generate comprehensive maps of PPIs, has enabled the development of a network-based view of biological processes (1, 2). Such networks are composed of ensembles of proteins that act together in a coordinated manner to execute a variety of essential biological processes, such as DNA replication, transcription, and signal transduction. In many cases, such networks are modular and contain highly connected proteins, termed “hub proteins,” that can regulate a given biological process by switching partners with high spatial and temporal resolution (3).
Many PPI networks are conserved over evolutionary time scales to promote a variety of biological processes in different organisms (4). One mechanism to maintain network structure and connectivity throughout evolution involves the coevolution of interacting proteins through coordinated changes in protein-protein interfaces (5). The coevolution of interacting proteins can form reproductive barriers between organisms as a result of hybrid network incompatibility, and thus can be an important driving force in promoting and fixing speciation. Currently, the study of coevolution of PPI networks is extremely challenging because of difficulties in identifying and characterizing coordinated sequence changes in network proteins during natural evolution. Even if such sequence changes are detected, their functional implications are difficult to predict. Thus, relatively little is known overall regarding the dynamics and functional importance of the coevolution of hub-partner interactions across different species.
In eukaryotes, DNA replication and repair processes are mediated by proliferating cell nuclear antigen (PCNA) through the recruitment of numerous DNA-modifying enzymes to the replication fork (6). PCNA is located at the heart of a complex PPI network, where it plays a key role in promoting DNA replication and repair processes (7). PCNA forms a sliding platform to enhance the processivity and catalytic activity of its various partners by tethering them to the DNA template. Interestingly, many PCNA partners compete for the same binding site on PCNA through a conserved binding motif (8). With our extensive knowledge of the molecular basis of PCNA-partner interactions (6, 9, 10), PCNA offers an excellent model system for the investigation of the coevolution of PPI networks. To date, however, little is known regarding the evolution of PCNA-partner interactions. For example, it is unclear whether PCNA coevolved with its partners to maintain interactions throughout evolution or whether these interactions are completely conserved. Such coevolution could lead to increased PCNA-binding specificity to cognate partners and reduced binding to distantly related partners, resulting in a lack of function of interspecies hybrids of PCNA-partner interaction networks.
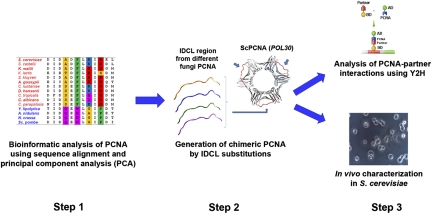
Here, we used bioinformatics and experimental tools to examine the coevolution of PCNA-partner interaction networks in fungal species spanning ∼300 million years of evolution (11). We performed sequence analysis of the most prominent PCNA-binding site in different fungal species to guide the generation of a series of interspecies chimeric PCNA variants. These variants were examined for PCNA-partner interactions with several partners from Schizosaccharomyces pombe, Saccharomyces cerevisiae, and Yarrowia lipolytica, as well as for in vivo functional complementation in S. cerevisiae (Fig. 1). Our analysis indicates the dramatic divergence of these networks in fungi into two distinct groups attributable to coevolution of PCNA-partner interactions. We found that such coevolution leads to a lack of function of heterologous PCNA in promoting DNA replication and repair in S. cerevisiae. These results indicate that the coevolution of PPI networks can lead to noncompatibility of hybrid networks, and thus promotes functional barriers between species.
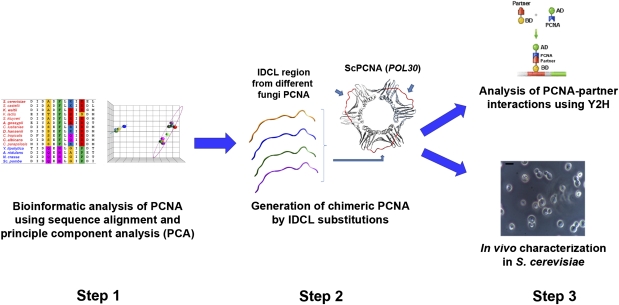
Fig. 1.
Bioinformatics and experimental workflow used for analysis of the coevolution of PCNA-partner interactions. Changes in the IDCL region of PCNA are analyzed by sequence alignment and PCA (step 1). Next, different chimeric PCNA variants are generated by substitution of the native IDCL region with IDCLs from different species (step 2). Such variants are characterized for PCNA-partner interactions and for in vivo function in S. cerevisiae as a model organism (step 3). Bar indicates a size of 5 microns.
Results
Combined Bioinformatics and Experimental Approach.
To examine the coevolution of PCNA-partner interactions, we focused on the PCNA interdomain connecting loop (IDCL), a region that is known to be involved in the binding of a large number of partners in different eukaryotes (6, 9, 10). We first performed bioinformatics analysis of the IDCL sequence of PCNA from diverse fungal species to detect any significant sequence changes that occurred during natural evolution (Fig. 1, step 1). Guided by this analysis, we generated chimeric PCNA variants containing different natural IDCL sequences grafted onto three PCNA backbones from S. cerevisiae, Y. lipolytica, or S. pombe (Fig. 1, step 2). Such an approach enabled an examination of the evolution of IDCL-mediated PCNA-partner interactions while minimizing the effects of amino acid substitution on the folding and stability of PCNA (10). Next, the chimeric variants were analyzed for binding to several PCNA partners from S. cerevisiae, Y. lipolytica, and S. pombe to detect changes in binding to cognate and distantly related partners. In parallel, the chimeric PCNA variants were examined for their in vivo activity in S. cerevisiae (Fig. 1, step 3) to determine whether changes in IDCL sequences have functional consequences on PCNA-mediated DNA replication and repair. In addition, we cloned several WT PCNA variants from different fungal species and examined their in vivo activity in S. cerevisiae and binding to cognate and distantly related partners. Overall, this approach enables the examination of both the molecular basis and the functional implications of the coevolution of PCNA-partner interactions. The generation of several chimeric and WT PCNAs to probe the fungal phylogenetic tree systematically enabled us to monitor the dynamics of coevolution of PCNA-partner interactions.
Bioinformatics Analysis of PCNA and Its Partners in Different Fungal Species.
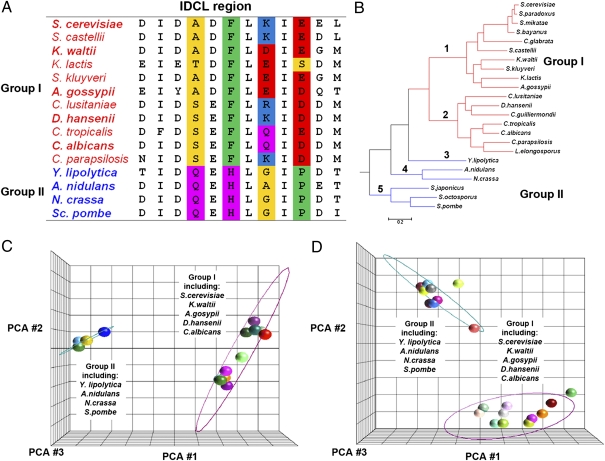
To study the evolution of PCNA-partner interactions in fungi, we first performed bioinformatics analysis of PCNA IDCL sequences. Sequence alignment of the IDCL sequences from the 23 fungal species for which a well-annotated phylogenetic tree exists (11) indicated significant divergence of these sequences (Fig. 2). We identified changes in four residues that are partially conserved within branches 1 and 2 of the fungal tree but are significantly different in branches 3–5 (Fig. 2 A and B). Next, we performed principal component analysis (PCA) of the different IDCL sequences and observed a sharp segregation of these sequences into two groups, where one group contains species found on branches 1 and 2 (group I) and the other group contains species found on branches 3–5 (group II, Fig. 2 B and C). Extending the PCA analysis to 53 PCNA IDCL sequences (Fig. S1) from different species retrieved from the fungal sequence database (http://www.yeastgenome.org/) revealed segregation into the same two groups (Fig. 2D). Finally, phylogenetic tree analysis of full-length PCNA, including the IDCL sequence, from 23 fungal species (11) agrees with the bioinformatics analysis of the IDCL region, indicating separation of the entire pool of PCNA sequences into two main branches comprising branches 1 and 2 and branches 3–5, respectively (Fig. S2). In addition, the C-terminal region of PCNA was previously shown to be important in mediating the interaction of PCNA with several partners essential for DNA replication (12). Sequence alignment of the C-terminal region of PCNA showed division into the same two groups as observed for the IDCL sequences, indicating coevolution of these two PCNA regions (Fig. S3).
Fig. 2.
Bioinformatics analysis of PCNA IDCL sequences across different species indicates divergence into two groups. (A) Sequence alignment of the IDCL region indicates changes mainly in four amino acids (highlighted in color) that can be used to separate the sequences into two groups (group I and group II; highlighted in red and blue, respectively). (B) Species phylogenetic tree containing 23 well-annotated fungal species (11). Group I and group II are highlighted in red and blue, respectively. (C) PCA of PCNA IDCL sequences from 23 species, including 15 unique sequences, indicates divergence into two groups; representatives of group I and group II are highlighted. The ellipse was generated by measuring 3 SDs around the centroid of each group, using Partek. (D) Similar analysis performed on IDCL sequences from 53 species, including 21 unique sequences (Fig. S1) found on BLAST analysis of fungal sequences (http://www.yeastgenome.org/). PCA was performed using the Jalview and Partek programs. The color scheme of amino acids in A is according to Lesk (28). Small nonpolar residues (G, A, S, T) are highlighted in yellow, hydrophobic residues (C, V, I, L, P, F, Y, M, W) are highlighted in green, polar residues (N, Q, H) are highlighted in magenta, negatively charged residues (D, E) are highlighted in red, and positively charged residues (K, R) are highlighted in blue.
It was previously shown, using several experimental approaches, that PCNA interacts with many different partners (Saccharomyces Genome Database, http://www.yeastgenome.org/). Analysis of PCNA partners indicates that the majority contain a conserved binding motif termed the “PCNA-interacting protein” (PIP) box, usually located at the N- or C-terminal region of the partner (6, 8). The PIP box interacts specifically with the IDCL region of PCNA and contains a clear QXXL/VXXFF signature (6, 8). To examine whether the PIP sequences of the different PCNA partners divide into the same two groups as observed for PCNA, we performed bioinformatics analysis of different PCNA partner sequences from the 53 fungal species listed in Fig. S1. These fungal species were assigned to group I or group II based on a clear division of the PCNA IDCL sequences (Fig. S1). In our analysis, we discarded all partner sequences in which the PIP box is not conserved, and thus could not be clearly identified or contained gaps. The PIP alignment revealed a divergence of these sequences into the same two groups as noted for PCNA. We observed divergence of the PIP sequences of several PCNA partners, including Pol32, Rad27, Ung1, Cdc1, Msh6, and Msh3 (Figs. S4–S6). Such separation was less clear than the separation between the PCNA IDCL sequences (Fig. 2), indicating the flexibility of PCNA-partner interactions. Interestingly, the PIP region could not be identified in several group II PCNA partners, indicating a lack of interaction between these partners and PCNA through the IDCL region (Fig. S7).
Generation and Characterization of Chimeric and WT PCNA Variants.
To study the effects of the dramatic changes in natural IDCL sequences on in vivo PCNA activity and PCNA-partner interactions, we generated and examined eight different chimeric PCNA variants. DNA encoding chimeric PCNAs was generated by overlapping PCR to incorporate the IDCL PCNA region from Kluyveromyces waltti, Ashbya gossipy, Debaryomyces hansenii, and Candida albicans (group I, Fig. 2) and from Y. lipolytica, Aspergillus nidulans, Neurospora crassa, and S. pombe (group II, Fig. 2) into the S. cerevisiae PCNA (ScPCNA or POL30) backbone (Fig. 1, step 2). In addition, we cloned and examined six different WT PCNAs, including that from K. waltti (KwPCNA), A. gossipy (AgPCNA), and D. hansenii (DhPCNA), all belonging to group I, and from Y. lipolytica (YlPCNA), N. crassa (NcPCNA), and S. pombe (SpPCNA or PCN1), all belonging to group II (Fig. 2).
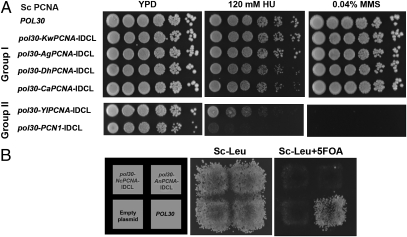
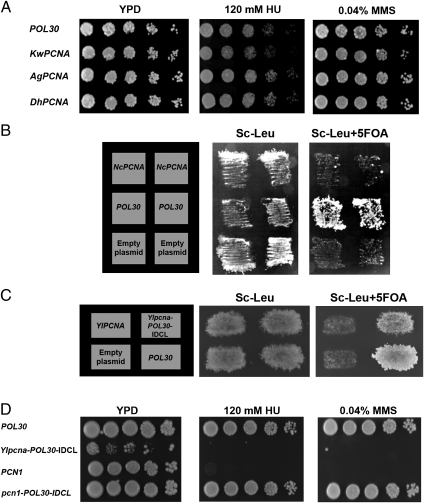
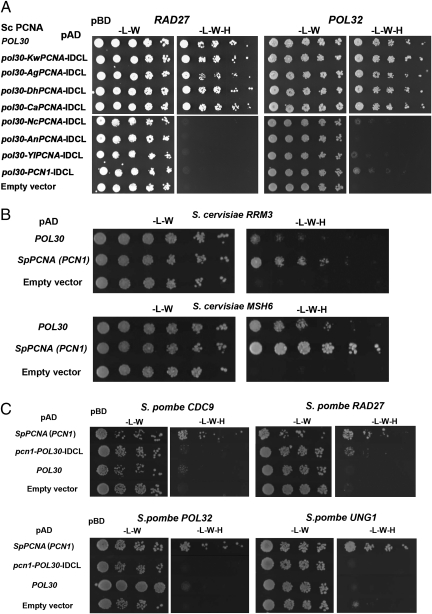
We first examined the in vivo function of the different chimeric and WT PCNAs in S. cerevisiae by generating yeast haploid strains containing chimeric PCNA as the sole source of PCNA in the cell. To generate such haploid strains, we used a plasmid shuffling approach utilizing yeast strains containing WT ScPCNA expressed from a URA3 plasmid and the chimeric PCNA variants expressed from a LEU2 plasmid. On selection for loss of ScPCNA using 5-fluoroorotic acid (5-FOA), a haploid strain containing the chimeric variant as the sole source of PCNA was generated (details are provided in Methods). In all cases, the PCNA variants were expressed from the native ScPCNA promoter to obtain similar expression levels. Our analysis indicated that strains containing chimeric PCNA with group I IDCL sequences exhibited no growth sensitivity in the presence of DNA-damaging agents, such as hydroxyurea (HU) or methymethanesulfonate (MMS), drugs that cause global replication stress and DNA alkylation, respectively (Fig. 3A). In contrast, strains containing chimeric PCNA with the Y. lipolytica or S. pombe IDCL sequence (group II, Fig. 2) exhibited increased sensitivity to HU and MMS, relative to the WT ScPCNA strain (Fig. 3A). Moreover, we found that strains containing the chimeric PCNA variants with the A. nidulans or N. crassa IDCL sequence were nonviable, indicating the inability of these chimeric PCNAs to support essential DNA replication processes in S. cerevisiae (Fig. 3B). In accordance with these results, strains containing KwPCNA, AgPCNA, or DhPCNA, all belonging to group I (Fig. 2), exhibited no growth sensitivity in the presence of HU or MMS (Fig. 4A). In addition, we observed that the strain containing SpPCNA PCN1 is highly sensitive to DNA-damaging agents, whereas strains containing NcPCNA or YlPCNA are nonviable (Fig. 4 B and C). These results highlight the functional incompatibility of group II PCNA in S. cerevisiae.
Fig. 3.
In vivo phenotypic analysis of the chimeric S. cerevisiae PCNA variants from group I and group II species. (A) Sensitivity of yeast strains containing chimeric PCNA variants as the sole source of PCNA to DNA-damaging agents, including HU (Center) and MMS (Right). (B) Nonviable strains containing chimeric ScPCNA with the N. crassa or A. nidulans IDCL sequence. Yeast strains containing ScPCNA expressed from a URA3 plasmid and chimeric Scpcna genes expressed from a LEU2 plasmid are viable (Center). On selection for loss of WT PCNA using 5-FOA, chimeric PCNA strains are deemed nonviable (Right).
Fig. 4.
In vivo phenotypic analysis of WT PCNAs from group I and group II species. (A) HU (Center) and MMS (Right) sensitivity of yeast strains containing the ScPCNA, KwPCNA, AgPCNA, or DhPCNA variants as the sole source of PCNA. (B) Nonviable strains containing NcPCNA. (C) Nonviability of the S. cerevisiae strain containing YlPCNA is suppressed by the replacement of the native IDCL region with the ScPCNA (POL30) IDCL region (Ylpcna-POL30-IDCL), indicating the importance of this region in facilitating strain survival (the generation of S. cerevisiae strains containing PCNA variants as a sole source of PCNA in the cell was performed as described in Fig. 3 and in the main text). (D) HU (Center) and MMS (Right) sensitivity of strains containing the SpPCNA the chimeric Ylpcna-POL30-IDCL, or the chimeric pcn1-POL30-IDCL gene. The sensitivity of the SpPCNA-containing strain is suppressed by replacement of its native IDCL region with the POL30 IDCL region (pcn1-POL30-IDCL).
To examine whether the IDCL sequences significantly affect the in vivo function of group II WT PCNAs, we generated additional chimeric PCNA variants based on the backbone of PCNA from group II species yet containing the IDCL region from group I species. We specifically generated chimeric variants with a backbone of either YlPCNA or SpPCNA containing the IDCL of ScPCNA. We observed that chimeric YlPCNA or SpPCNA containing the ScPCNA IDCL region is viable or shows a lack of sensitivity to HU or MMS, respectively, indicating that the replacement of the IDCL leads to suppression of parental gene phenotypes (Fig. 4 C and D). These results further highlight the critical importance of the IDCL region in coordinating the binding of multiple partners and promoting in vivo DNA replication and repair in S. cerevisiae. Overall, the results obtained with the chimeric and WT PCNA variants indicate that the divergence of the PCNA and IDCL sequences to form two distinct groups leads to loss of function in S. cerevisiae (Figs. 3 and 4), highlighting the incompatibility of hybrid PCNA-partner interaction networks. Moreover, the groups of species diverged according to their distances in the phylogenetic tree (Fig. 2), indicating divergence from a common ancestor.
Analysis of PCNA Interactions with S. cerevisiae Partners.
To characterize changes in PCNA-partner interactions during fungal evolution, we examined the binding of chimeric and WT PCNA variants to several S. cerevisiae PCNA partners, including Pol32, Rad27, Rad30, Cdc9, Apn2, and Ung1, using a yeast two-hybrid approach (13). We observed that chimeric PCNAs containing IDCL sequences from group I species (Fig. 2) were able to bind the different partners in a manner similar to WT ScPCNA (Fig. 5A and Table 1). In contrast, chimeric PCNA containing the IDCL from group II species (Fig. 2) exhibited dramatically reduced binding to the different ScPCNA partners (Fig. 5A and Table 1). To verify that these chimeric PCNA variants are correctly translated and stable in S. cerevisiae, we established an ELISA to monitor the expression levels of the different PCNA variants in S. cerevisiae (Fig. S8). These experiments show that all chimeric PCNA variants containing the group II IDCL region are expressed at similar levels as WT ScPCNA (Fig. S8).
Fig. 5.
Representative yeast two-hybrid analysis of chimeric and WT PCNA variants interacting with group I and group II PCNA partners. (A) Representative analysis of the interaction of chimeric PCNA with S. cerevisiae RAD27 (Left) or POL32 (Right). A full-yeast two-hybrid analysis is provided in Table 1. (B) Representative analysis of the interactions of ScPCNA or SpPCNA with S. cerevisiae Rrm3 and Msh6. SpPCNA exhibits higher interaction levels with the two partners, relative to ScPCNA (results of a full-yeast two-hybrid analysis are provided in Table S1). (C) Yeast two-hybrid analysis of the SpPCNA, ScPCNA (POL30), and chimeric SpPCNA variants interacting with CDC9, RAD27, POL32, and UNG1 orthologs in S. pombe. Interactions can be detected in most cases only with SpPCNA but not with ScPCNA or the chimeric SpPCNA variants containing the ScPCNA IDCL region (pcn1-POL30-IDCL; Table 2).
Table 1.
Y2H analysis of chimeric PCNA-partner interactions
|
S. cerevisiae PCNA partners |
||||||
| Sc PCNA (POL30) | Pol32 | Rad27 | Ung1 | Cdc9 | Rad30 | Apn2 |
| POL30 | ++++ | ++++ | +++ | ++++ | ++++ | +++ |
| Pol30-KwPCNA-IDCL | ++++ | ++++ | +++ | ++++ | ++++ | +++ |
| pol30-AgPCNA-IDCL | ++++ | ++++ | +++ | ++++ | ++++ | +++ |
| pol30-DhPCNA-IDCL | ++++ | ++++ | +++ | ++++ | ++++ | +++ |
| pol30-CaPCNA-IDCL | ++++ | ++++ | +++ | ++++ | ++++ | +++ |
| pol30-NcPCNA-IDCL | + | − | ND | + | − | + |
| pol30-AnPCNA-IDCL | − | − | − | − | − | − |
| pol30-YlPCNA-IDCL | + | − | − | + | − | + |
| pol30-SpPCNA-IDCL | + | − | +++ | + | + | ++ |
| KwPCNA | ++++ | ++++ | +++ | ++++ | ND | ND |
| AgPCNA | ++++ | ++++ | +++ | ++++ | ND | ND |
| DhPCNA | ++ | ++ | ++ | ++ | ND | ND |
POL30 and chimeric pol30 variants were fused to the pAD, and the partners were fused to the pBD. Each yeast two hybrid (Y2H) strain was spotted in five dilutions (e.g, Fig. 5), and the interactions were scored according to the growth of the different dilutions on selective plates lacking histidine (−, no growth; +, growth observed at first serial dilution; ++, growth observed at second serial dilution; +++, growth observed at third serial dilution; ++++, growth observed at fourth serial dilution). ND, not determined.
Next, we examined the binding of WT PCNAs from group I and group II to S. cerevisiae partners. We found that KwPCNA and AgPCNA, belonging to group I (Fig. 2), were able to bind the different partners in a manner similar to WT ScPCNA, whereas DhPCNA exhibits a reduced level of interaction that is, nonetheless, still much higher than the background level (Table 1). Surprisingly, we found that SpPCNA or YlPCNA exhibited high levels of these interactions that, in some cases, exceeded the interaction affinity of ScPCNA to its cognate partners (Fig. 5B and Table S1). The low level of functional complementation of SpPCNA or YlPCNA in S. cerevisiae indicates that these interactions are, however, nonproductive in terms of promoting DNA replication (Fig. 4 C and D). These interactions could be mediated by other regions of SpPCNA or YlPCNA that are distal from the IDCL. Unfortunately, because of the lack of structural information on SpPCNA- or YlPCNA-partner interactions, it is difficult to understand the molecular basis for such interactions and to identify those PCNA regions that directly interact with the different partners. The increased levels of SpPCNA- or YlPCNA-partner interactions could lead to the low in vivo functionality of these PCNA orthologs in S. cerevisiae (Fig. 4). We have previously shown that a fine balance between the different PCNA-partner interaction affinities is essential for promoting DNA replication and repair (10); thus, any alteration of such balance, including minor increases in affinity, can lead to a loss of function in S. cerevisiae.
Analysis of PCNA Interactions with S. pombe or Y. lipolytica Partners.
To examine PCNA-partner interactions in group II species, we cloned several PCNA partners from the Y. lipolytica and S. pombe genomes, including POL32, RAD27, CDC9, RRM3, and UNG1 orthologs. We first examined the binding of YlPCNA or SpPCNA to the different partners and verified that, indeed, each variant can bind the majority of its cognate or closely related partners (Fig. 5C and Table 2). In contrast, we observed that the interaction affinities of chimeric YlPCNA or SpPCNA containing the ScPCNA IDCL region to the same partners were significantly reduced, indicating a lack of compatibility between the ScPCNA IDCL region and group II partners (Fig. 5C and Table 2). This observation was further strengthened by the lack of binding of the WT ScPCNA, KwPCNA, AgPCNA, or DhPCNA to group II partners (Table 2).
Table 2.
Y2H analysis of fungal PCNA interactions with S. pombe or Y. lipolytica PCNA partners
|
S. pombe PCNA partners |
Y. lipolytica PCNA partners |
|||||||
| Chimeric PCNA | Pol32 | Rad27 | Rrm3 | Ung1 | Pol32 | Rad27 | Cdc9 | Ung1 |
| POL30 | − | − | − | − | − | − | − | − |
| KwPCNA | − | − | − | − | ND | ND | ND | ND |
| AgPCNA | − | − | − | − | ND | ND | ND | ND |
| DhPCNA | − | − | − | − | ND | ND | ND | ND |
| PCN1 | ++ | + | ++ | + | ++ | ++ | ++ | ++ |
| pcn1-POL30-IDCL | − | − | − | − | − | + | − | − |
| YlPCNA | + | + | + | ++ | − | + | + | + |
| Ylpcna-POL30-IDCL | − | − | − | − | − | − | − | − |
POL30 and chimeric pol30 variants were fused to the pAD, and the partners were fused to the pBD. The interactions were scored according to the growth of the different dilutions on selective plates lacking histidine (−, no growth; +, growth observed at first serial dilution; ++, growth observed at second serial dilution; +++, growth observed at third serial dilution; ++++, growth observed at fourth serial dilution). ND, not determined.
Laboratory Evolution of YlPCNA for Functional Complementation of ScPCNA in S. cerevisiae.
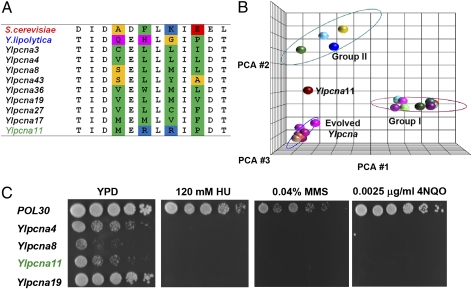
Our bioinformatics analysis indicates that the classification of IDCL sequences into two groups is mainly attributed to four positions within the IDCL sequence that are differentially conserved within each group (Fig. 2A). To examine whether the group I IDCL sequence is the only natural solution for promoting DNA replication and repair in S. cerevisiae, we performed a laboratory evolution experiment. In recent years, directed protein evolution approaches have proven to be highly effective for generating proteins with unique functions, including binding and catalysis (14–16). Using laboratory evolution, we randomly mutated the four positions in the IDCL region of YlPCNA (Fig. 2) and selected ∼2,500 random Ylpcna mutants for viability as a sole source of PCNA in S. cerevisiae (details are provided in Methods). Following screening, we identified 9 unique Ylpcna mutants that could complement the deletion of ScPCNA in S. cerevisiae, reflecting a frequency of ∼0.4% of positive clones in the library (Fig. S9). Sequence analysis of the different mutants showed an enrichment of different combinations of hydrophobic residues (i.e., L, V, I, F) in the targeted positions (Fig. 6A). PCA analysis indicates that these sequences form a distinct group that is different from the natural group I and group II IDCL sequences (Fig. 6B). The isolation of such sequences at a relatively high frequency indicates the existence of many possible solutions to promote DNA replication and repair in S. cerevisiae. In contrast to the selected hydrophobic IDCL mutants, we found that 1 mutant, Ylpcna11, contains the MRRP hydrophilic sequence in its IDCL region (Fig. 6A). Such a sequence combines a proline residue present in YlPCNA and a positively charged arginine residue, similar to the lysine found in ScPCNA, at the respective position (Fig. 6A). Given the small number of mutants screened from the library, it is likely that many more sequences similar to Ylpcna11 can be isolated from the naive Ylpcna library. In vivo functional analysis of selected Ylpcna mutants in S. cerevisiae indicated high levels of sensitivity to HU or MMS (Fig. 6C), similar to the sensitivity observed with chimeric YlPCNA containing the ScPCNA IDCL region (Fig. 4). These results suggest that the IDCL region must coevolve with other regions of PCNA [e.g., C-terminal region (Fig. S3)] to achieve optimal in vivo function. Finally, we examined the binding of selected mutants to an YlPCNA, SpPCNA, or ScPCNA partner. Interestingly, we observed that YlPCNA11 is the only mutant that maintains interactions with partners from Y. lipolytica and S. pombe (Fig. S10 and Table S2) and promotes DNA replication and repair in S. cerevisiae.
Fig. 6.
In vitro evolution of Y. lipolytica PCNA for complementation of ScPCNA in S. cerevisiae. (A) Sequence alignment of 9 unique mutants identified following selection for viability. The mutated positions are mostly hydrophobic and are highlighted in color (using the color scheme described in the legend for Fig. 2). (B) PCA analysis of the 9 unique sequences of the evolved clones, together with the 23 natural IDCL sequences (described in Fig. 2). The analysis indicates that the evolved Ylpcna mutants fall into a unique group that is different from the two natural groups, except for YlPCNA11, which is significantly different in its sequence. (C) HU, MMS, and 4-nitroquinoline-N-oxide (4NQO) sensitivity of the evolved Ylpcna mutants.
Discussion
In this work, we used the PCNA-partner interaction network as a model system to examine the coevolution of complex PPI networks in fungi. We used a combination of bioinformatics and experimental approaches to detect changes in the PCNA sequence throughout fungal evolution and examined the effects of such changes on PCNA-partner interactions and on in vivo PCNA function in S. cerevisiae. The results obtained with the chimeric variants revealed the divergence of PCNA-partner interaction networks into two distinct groups as a result of the coevolution of IDCL-mediated interactions across yeast species. Our results show that the coevolution of PCNA-partner interactions can lead to the lack of function in a hybrid PCNA network. The classic Dobzhansky–Muller model proposes that hybrid incompatibility, important for speciation, evolves as a result of interactions of genes that have diverged in the respective hybridizing species (17). Such divergence can evolve if populations of fungi are isolated (e.g., allopartic populations) as a result of geographical changes. The isolated fungal populations can then undergo genotypic and phenotypic divergence, leading to sequence and functional changes in PCNA and its partners. Divergence can also result from adaptation to different environmental conditions, leading to the fixation of different types of PCNA and partner sequences. Once incompatibility evolves, it can serve as a reproductive barrier to stop gene flow between populations, thus leading to speciation. According to this model, it is possible that diverged PCNA-partner networks play a role in initiating speciation. Still, the divergence of the PCNA-partner interaction network is unlikely to be a key factor in promoting fungal speciation because such divergence probably occurred only once during 300 million years of yeast evolution.
Sequence analysis of PCNA partners from many fungal species can provide insight into the possible dynamics of the dramatic sequence changes between the two groups of fungi species. Analysis of PCNA partners shows that in several cases, the PIP region is deleted/mutated (Fig. S7), indicating that during evolution, a substantial loss or gain of PCNA-partner interactions (mediated through the IDCL-PIP interaction) took place. We found lower number of partners containing the PIP sequence in group II species (Fig. S7), highlighting the flexibility of the PCNA-partner interaction network through evolution. However, we cannot exclude the possibility that partners lacking the PIP box interact with PCNA through other regions or interact directly with the DNA template. Overall, the flexibility and versatility of this network in fungal species offers a possible gradual mode of coadaptation of PCNA and its partners through evolution.
To date, the coevolution of interacting proteins has been extensively examined using two main approaches. The first approach identifies correlating site-specific mutations in interacting protein interfaces to enable the maintenance of PPIs over evolution (18). The second approach that is broadly used relies on the similarity of phylogenetic trees of the interacting protein family by comparing pair-wise distance matrices derived from amino acid sequence alignments (i.e., the “mirror-tree” approach) (19–21). Both of these approaches are based on bioinformatics analysis and were performed in many cases on large numbers of proteins with the intent of predicting probable PPIs in other species. In contrast, very little experimental work addressing the molecular basis and functional implications of PPI coevolution has been performed. Our combined bioinformatics and experimental approaches can thus provide a unique means for the detection and characterization of the coevolution of such networks. Here, this approach enabled detection of the coevolution of transient PCNA-partner interactions and its functional consequences with no need for identifying correlated mutations in both PCNA and its partners. Further application of directed evolution indicated that the divergence of the PCNA IDCL sequence from a common ancestor could have taken place via several routes (Fig. 6). The findings also highlight the flexibility and evolvability of the IDCL region for binding multiple partners and for promoting DNA replication and repair.
Is it likely that similar divergence in other PPI networks can be found? Residues that are differentially conserved in protein subfamilies were previously identified in many different proteins (22). Such residues, termed “specificity-determining residues,” were shown to be involved in ligand binding and catalytic activities (23). In some instances, mainly in the case of enzyme specificity, the importance of such residues was experimentally validated (24). It was also shown that such changes can take place simultaneously, leading to a cluster of differentially conserved, coevolving residues that diverged over evolution (25). Thus, PCNA may be only one example of a hub protein that diverged into distinct subtypes, with such changes being prevalent in other protein families. However, an experimental approach must be taken to examine and characterize the roles of differentially conserved residues in the coevolution of other proteins.
In summary, we have shown that the PCNA-partner interaction network diverged into two groups because of significant coevolution of the interactions. Our data indicate that such coevolution can lead to a loss of function of interspecies hybrid networks. Overall, our combined bioinformatics and experimental approach may pave the way for examination of the coevolution of a variety of other PPI networks that mediate diverse biological processes, including signal transduction and gene transcription.
Methods
Plasmids.
For in vivo testing of PCNA mutants, the 200-bp PCNA-promoter region and the 300-bp PCNA-terminator region were amplified from genomic DNA using specific primers and cloned into the pRS315 and pRS316 centromeric plasmids using NotI and SpeI sites and HindIII and XhoI sites, respectively. The resulting pRS315-proterm and pRS316-proterm plasmids were used to introduce the WT and chimeric PCNA genes by homologous recombination using primers forward (fr)-pRS-PCNA and reverse (rev)-pRS-PCNA (Table S3). The chimeric PCNA variants were generated by amplifying POL30, S. pombe PCNA, or YlPCNA to yield two fragments with complementary IDCL regions. These fragments were assembled by PCR and amplified using the fr-pRS-PCNA and rev-pRS-PCNA primers to facilitate homologous recombination. For the yeast two-hybrid assay, the DNA-activating domain (pAD)-GAL4 and DNA-binding domain (pBD)-GAL4 plasmids (Stratagene) were used for the cloning of WT and chimeric PCNA and the different partners, respectively. pAD-PCNA and pAD-PCNA-chimera were generated by amplification of the WT and mutant PCNA genes using primers fr-pAD-PCNA and rev-pAD-PCNA from the pRS library, followed by homologous recombination in yeast. The pBD-partners collection was generated by sequence-specific amplification of each partner from yeast genomic DNA. Cloning was facilitated by homologous recombination of the different inserts using common overhang sequences of 50 bp. Y. lipolytica and S. pombe PCNA and PCNA partners were amplified from genomic DNA and cDNA for homologous recombination. Total RNA was isolated (EZ RNA Kit; Biological Industries), followed by RT-PCR (SuperScriptII; Invitrogen) to generate cDNA for each gene.
YlPCNA Library Generation.
Positions Q123, H125, G127, and P129 of YlPCNA were fully randomized by two-fragment overlapping PCR using plasmid pRS-YlPCNA as a template. The two pcna gene fragments were amplified using two sets of primers (fr-lipolytica-pcna/pRS and rev-Libx4, rev-lipolytica-pcna/pRS and fr-Libx4). Fragments were then assembled and amplified with a set of primers containing plasmid pRS complementary overhangs. The library was transformed into the S. cerevisiae haploid strain and grown on −Ura/−Leu plates. Clones were manually patched, replicated to 5-FOA plates, and screened for viability. Viable clones were sequenced and retransformed into the haploid strain for confirmation.
Yeast Two-Hybrid Analysis.
Yeast two-hybrid analysis was performed using the Yeast Two-Hybrid Phagemid vector kit (Stratagene), following the manufacturer's instructions. Selected PCNA mutants and partners were cloned in-frame with the GAL4 pBD and pAD, respectively, as described above. The pJ69-4A or YRG2 (Stratagene) host strain (26) was cotransformed with either the pAD-PCNA or pAD-PCNA chimera plasmids and a pBD-partner in all possible combinations using the lithium acetate (LiAc) method. Single transformants were grown on SC-Leu-Trp plates, and single colonies were grown in liquid SC-Leu-Trp to OD600 of 10 nm, washed, and diluted to an initial OD600 of 0.3. A series of serial dilutions was conducted, and the cells were spotted on selective Synthetic Complete (SC)-Leu-Trp-His plates. In all cases, a comparison of growth among cells transformed with pAD-PCNA-chimera, pAD-PCNA-WT, and empty pAD plasmids was performed and interaction affinity was scored based on yeast growth.
ELISA for Assessing Chimeric PCNA Expression Levels.
Yeast cell extracts were generated from 0.5-L logarithmic cultures using conventional methods. Briefly, cell pellets were resuspended and vigorously shaken in cell lytic solution (Sigma) supplemented with protease inhibitors (1:100; Sigma) and glass beads. Following centrifugation, the clear extracts were collected and total protein concentration was determined using the BCA method (Pierce). ELISA plates coated with mouse α-pAD antibodies (1:50; Santa Cruz Biotechnology) were incubated with 100 μL of yeast cell extract at a protein concentration of 3 mg/mL for 2 h at room temperature (RT). Following three washing steps with PBS supplemented with 0.05% Tween-80 (PBST), the plate wells were incubated with α-PCNA antibodies (1:250; Adar Biotech) for 1 h at RT. Plates were then washed three times with PBST and incubated with HRP-conjugated goat α-rabbit antibodies (1:2,000; Jackson). Finally, HRP-chromogenic TMB substrate solution (Dako) was added. The reaction was stopped by the addition of 100 μL of 1 M sulfuric acid and recorded at 450 nm using a Tecan Infinite M200 plate reader. Values represent averages of at least three independent repeats.
In Vivo Characterization of Unique PCNA Mutants.
Unique haploid PCNA mutant strains were generated using the plasmid shuffling method, as previously described (10). To characterize the in vivo phenotype of the mutants in the presence of DNA-damaging agents, cells were grown to OD600 of 10 serially diluted, and spotted onto plates containing 120 mM HU (Toronto Research Chemicals) or 0.04% MMS (Sigma). Plates were incubated for 3 d at 30 °C and shielded from light.
Bioinformatics Analysis.
Fungal PCNA orthologs were retrieved through a BLAST search of fungal sequences (http://www.yeastgenome.org/cgi-bin/blast-fungal.pl), using S. cerevisiae PCNA as a query. Fungal ortholog groups for each PCNA partner were obtained from the KEGG orthology database (http://www.genome.jp/kegg/) and aligned using ClustlW (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The PIP box and the preceding and following amino acids were extracted from the alignments. Sequences containing gaps in their PIP sequence were removed from the alignments. Fifty-three orthologs were chosen by their score and an e−10 threshold. Multiple sequence alignment and PCNA and IDCL sequence tree construction were performed using MEGA software (27). An established phylogenetic tree of 23 fungal species was adopted from the fungal orthogroups repository (http://www.broadinstitute.org/regev/orthogroups/). Their corresponding PCNA sequences were adopted from this database (11). PCA analysis of the PCNA orthologs was performed using Jalview 2.6 software (http://www.jalview.org/) and Partek Inc.
Supplementary Material
Acknowledgments
We thank Dan Mishmar, Daniel Dovrat, and Mehtap Abu-Qarn for critical reading of the manuscript and other colleagues for discussion. A.A. is supported by European Research Council “Ideas Program” Grant 201177.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 2200.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1108633109/-/DCSupplemental.
References
- 1.Bork P, et al. Protein interaction networks from yeast to human. Curr Opin Struct Biol. 2004;14:292–299. doi: 10.1016/j.sbi.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Krogan NJ, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 3.Han JD, et al. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature. 2004;430:88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- 4.Sharan R, et al. Conserved patterns of protein interaction in multiple species. Proc Natl Acad Sci USA. 2005;102:1974–1979. doi: 10.1073/pnas.0409522102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pazos F, Valencia A. Protein co-evolution, co-adaptation and interactions. EMBO J. 2008;27:2648–2655. doi: 10.1038/emboj.2008.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 7.Warbrick E. The puzzle of PCNA's many partners. Bioessays. 2000;22:997–1006. doi: 10.1002/1521-1878(200011)22:11<997::AID-BIES6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 8.Maga G, Hubscher U. Proliferating cell nuclear antigen (PCNA): A dancer with many partners. J Cell Sci. 2003;116:3051–3060. doi: 10.1242/jcs.00653. [DOI] [PubMed] [Google Scholar]
- 9.Krishna TS, Kong XP, Gary S, Burgers PM, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 10.Fridman Y, et al. Subtle alterations in PCNA-partner interactions severely impair DNA replication and repair. PLoS Biol. 2010;8:e1000507. doi: 10.1371/journal.pbio.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wapinski I, Pfeffer A, Friedman N, Regev A. Natural history and evolutionary principles of gene duplication in fungi. Nature. 2007;449:54–61. doi: 10.1038/nature06107. [DOI] [PubMed] [Google Scholar]
- 12.Gomes XV, Burgers PM. Two modes of FEN1 binding to PCNA regulated by DNA. EMBO J. 2000;19:3811–3821. doi: 10.1093/emboj/19.14.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fields S, Sternglanz R. The two-hybrid system: An assay for protein-protein interactions. Trends Genet. 1994;10:286–292. doi: 10.1016/0168-9525(90)90012-u. [DOI] [PubMed] [Google Scholar]
- 14.Aharoni A, Griffiths AD, Tawfik DS. High-throughput screens and selections of enzyme-encoding genes. Curr Opin Chem Biol. 2005;9:210–216. doi: 10.1016/j.cbpa.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Arnold FH, Wintrode PL, Miyazaki K, Gershenson A. How enzymes adapt: Lessons from directed evolution. Trends Biochem Sci. 2001;26(2):100–106. doi: 10.1016/s0968-0004(00)01755-2. [DOI] [PubMed] [Google Scholar]
- 16.Tao H, Cornish VW. Milestones in directed enzyme evolution. Curr Opin Chem Biol. 2002;6:858–864. doi: 10.1016/s1367-5931(02)00396-4. [DOI] [PubMed] [Google Scholar]
- 17.Wu CI, Ting CT. Genes and speciation. Nat Rev Genet. 2004;5(2):114–122. doi: 10.1038/nrg1269. [DOI] [PubMed] [Google Scholar]
- 18.Pazos F, Valencia A. In silico two-hybrid system for the selection of physically interacting protein pairs. Proteins. 2002;47:219–227. doi: 10.1002/prot.10074. [DOI] [PubMed] [Google Scholar]
- 19.Goh CS, Cohen FE. Co-evolutionary analysis reveals insights into protein-protein interactions. J Mol Biol. 2002;324:177–192. doi: 10.1016/s0022-2836(02)01038-0. [DOI] [PubMed] [Google Scholar]
- 20.Pazos F, Ranea JA, Juan D, Sternberg MJ. Assessing protein co-evolution in the context of the tree of life assists in the prediction of the interactome. J Mol Biol. 2005;352:1002–1015. doi: 10.1016/j.jmb.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 21.Ramani AK, Marcotte EM. Exploiting the co-evolution of interacting proteins to discover interaction specificity. J Mol Biol. 2003;327:273–284. doi: 10.1016/s0022-2836(03)00114-1. [DOI] [PubMed] [Google Scholar]
- 22.Hannenhalli SS, Russell RB. Analysis and prediction of functional sub-types from protein sequence alignments. J Mol Biol. 2000;303:61–76. doi: 10.1006/jmbi.2000.4036. [DOI] [PubMed] [Google Scholar]
- 23.Rausell A, Juan D, Pazos F, Valencia A. Protein interactions and ligand binding: From protein subfamilies to functional specificity. Proc Natl Acad Sci USA. 2010;107:1995–2000. doi: 10.1073/pnas.0908044107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morillas M, et al. Identification of conserved amino acid residues in rat liver carnitine palmitoyltransferase I critical for malonyl-CoA inhibition. Mutation of methionine 593 abolishes malonyl-CoA inhibition. J Biol Chem. 2003;278:9058–9063. doi: 10.1074/jbc.M209999200. [DOI] [PubMed] [Google Scholar]
- 25.Gloor GB, Martin LC, Wahl LM, Dunn SD. Mutual information in protein multiple sequence alignments reveals two classes of coevolving positions. Biochemistry. 2005;44:7156–7165. doi: 10.1021/bi050293e. [DOI] [PubMed] [Google Scholar]
- 26.Chockalingam K, Chen Z, Katzenellenbogen JA, Zhao H. Directed evolution of specific receptor-ligand pairs for use in the creation of gene switches. Proc Natl Acad Sci USA. 2005;102:5691–5696. doi: 10.1073/pnas.0409206102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary Distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lesk A. Introduction to Bioinformatics. Oxford Univ Press, Oxford; 2008. [Google Scholar]