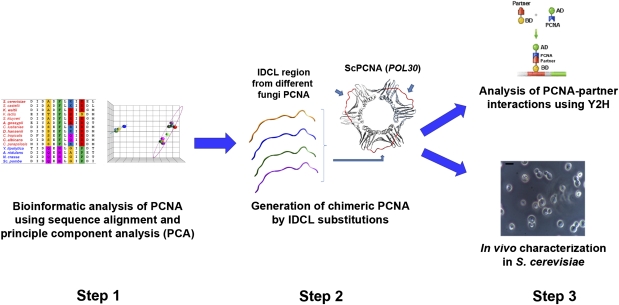
Fig. 1.
Bioinformatics and experimental workflow used for analysis of the coevolution of PCNA-partner interactions. Changes in the IDCL region of PCNA are analyzed by sequence alignment and PCA (step 1). Next, different chimeric PCNA variants are generated by substitution of the native IDCL region with IDCLs from different species (step 2). Such variants are characterized for PCNA-partner interactions and for in vivo function in S. cerevisiae as a model organism (step 3). Bar indicates a size of 5 microns.