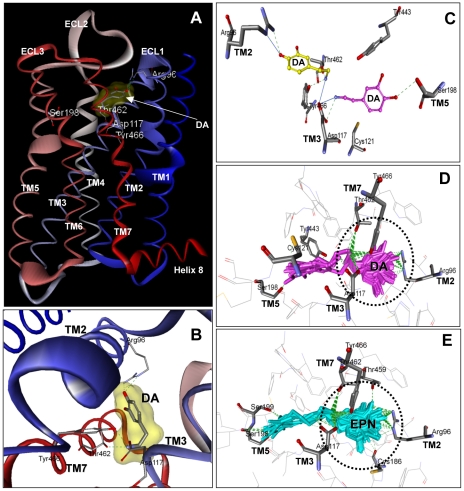
Figure 8. Homology modeling of SmGPR-3 and ligand docking.
(A) A homology model of SmGPR-3 with bound dopamine (DA) is shown. The model was generated using the β-2 adrenergic receptor (PDB Accession # 2rh1) as a structural template, as described in the Methods. The positions of the 7 predicted transmembrane (TM) helices and 3 extracellular loops (ECL) are marked. The additional intracellular helix at the C-terminal end (helix 8) is also shown. Amino acid residues that are predicted to interact with dopamine include: Arg 96 (R2.64), Asp117 (D3.32), Thr462 (T7.39) and Tyr466 (Y7.43). (B) Close-up of the predicted binding pocket showing the best docking pose of dopamine (DA) and the principal ligand binding residues. (C) Two different docking poses of dopamine (DA) are shown. Note that the position of the catechol ring in the two conformations is reversed. In the best scoring pose (yellow), the ring hydroxyl interacts with Arg96 (R2.64) near the extracellular junction of TM2, whereas in the other docking pose (magenta) the ring interacts with Ser198 (S5.42) of TM5 instead. In both cases the protonated amino end of dopamine is anchored to Asp117 (D3.32) of TM3. An overlay of potential docking poses of dopamine (DA, panel D) and the structurally related ligand, epinine (EPN, panel E) shows the majority of interactions occurring with TM2, 3 and 7 for both ligands. The core interacting residues are shown and predicted H-bonds are marked by broken green lines.