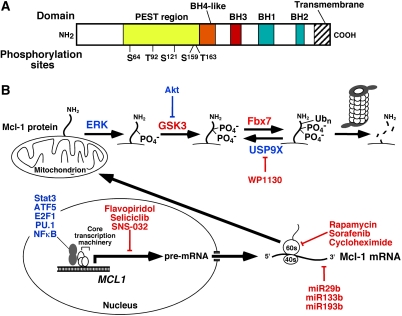
Figure 1.
Regulation of Mcl-1 levels. (A) Domain structure of Mcl-1. Shown are the four BH domains that Mcl-1 shares with Bcl-2 and other family members, as well as the localization of the proline/glutamic acid/serine/threonine-rich (PEST) region that contains many of the known Mcl-1 phosphorylation sites, including Ser 64 (Jun N-terminal kinase), Thr 92 (cyclin-dependent kinase 1), Ser 121 (extracellular signal-regulated kinase [ERK]), Ser 159 (glycogen synthase kinase-3 [GSK3] and PKCδ), and Thr 163 (ERK). (B) Transcriptional, translational, and post-translational regulation of Mcl-1 described in the text. Molecules depicted in blue increase Mcl-1 levels, whereas molecules rendered in red decrease Mcl-1 levels. In particular, ERK-mediated phosphorylation on Thr 163, which can stabilize Mcl-1 under certain circumstances, creates a recognition site for GSK3. GSK3-mediated phosphorylation of Mcl-1 on Ser 159 then creates the recognition site for the E3 ubiquitin ligase Fbx7, which generates Lys 48-linked ubiquitin chains that result in proteasome-mediated Mcl-1 degradation.