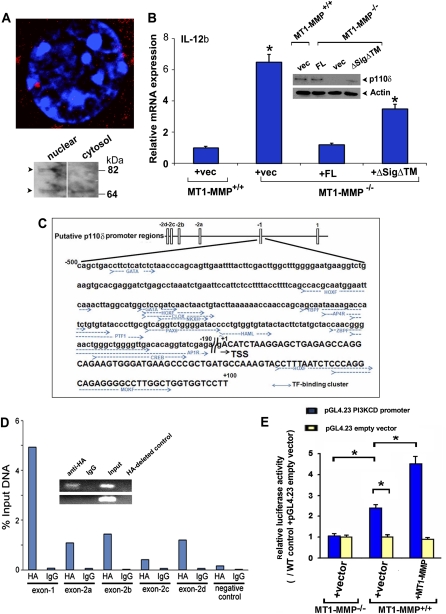
Figure 6.
MT1-MMP nuclear routing and interactions with the p110δ promoter. (A) Nuclear localization of ΔSigΔTM in transduced MT1-MMP−/− macrophages as assessed in confocal laser, Z-section micrographs or by Western blot analyses of the macrophage cytosolic and nuclear fractions. (B) IL-12b mRNA expression was determined in control vector transduced MT1-MMP+/+ macrophages or MT1-MMP−/− macrophages transduced with a control vector, full-length MT1-MMP (FL) or ΔSigΔTM following a 4-h incubation with LPS. In tandem, macrophage lysates were prepared from each sample, and p110δ protein levels were assessed by Western blot analysis. (C) Schematic representation of the p110δ promoter regions and the exon −1 region. Transcription factor (TF)-binding clusters are indicated in blue (Kok et al. 2009). (D) Wild-type or MT1-MMP-null macrophages (data not shown) were transduced with a control vector or HA-tagged MT1-MMP, and ChIP of PCR was performed with anti-HA antibody or control IgG. Results are presented as the mean of two experiments. (Inset) A representative image of semiquantitative PCR for the p110δ promoter (exon −1 region; top row) and negative control primers (+5 kb coding region primer; bottom row). Primers specific for the p110δ promoter were designed to amplify −358 to −19 of exon −1 (Kok et al. 2009). (E) Relative luciferase activity of the pGL4.23 and the pGL4.23 PI3KCD promoter in MT1-MMP−/− macrophages, MT1-MMP+/+ macrophages transduced with a control expression vector, or an MT1-MMP expression vector. Results are expressed as the mean ± SEM of five experiments; (*) P < 0.01; versus MT1-MMP+/+ + pGL4.23 empty vector + pcDNA 3.1 control vector.