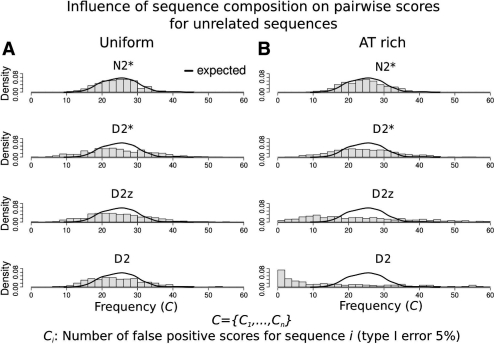
Fig. 2.
Influence of single sequences on pairwise scores. All pairwise scores for 500 sequences generated by the same model were calculated. Ci measures the number of sequence pairs for sequence Si among the highest 5% of all scores (high scoring pairs). Since all sequences were created using the same model, the distribution of C={C1,…, Ci} from alignment-free methods should be similar to the distribution of C obtained from a random scoring method (‘expected’, black line). A different distribution would indicate that the number of high scoring pairs is strongly dependent on the individual sequence, indicating that pairwise scores are dependent on the single sequence noise rather than on the similarity of the sequence pair. (A) Uniform nucleotide distribution, all methods show the expected behaviour. (B) AT-rich nucleotide distribution, D2 and D2z differ from the expected behaviour, showing that these pairwise scores are strongly influenced by the sequence composition.