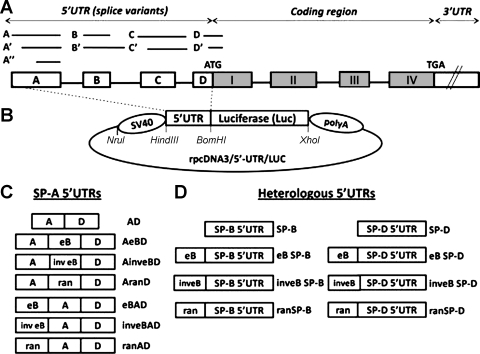
Fig. 1.
A: SP-A1 and SP-A2 common gene structure (not to scale). The 5′ untranslated region (5′UTR) exons A, B, C, and D differ in length, as represented by the lines and boxes. Alternative splicing of the 5′UTR exons gives rise to several SP-A1 and SP-A2 variants. All the SP-A2 mRNA variants described to date contain exon B (20). B: basic rpcDNA3/5′-UTR/LUC vector used to generate experimental plasmids. The rpcDNA3/5′-UTR/LUC was used to clone the experimental spliced 5′UTR sequences upstream of the luciferase reporter gene, as described in materials and methods. The SV40 promoter is located upstream of a 5′UTR sequence and a firefly luciferase cassette, followed by a polyadenylation sequence (polyA). The 5′UTR sequence is flanked by HindIII and BamHI restriction sites, and an XhoI site is located downstream of the luciferase coding sequence. C: diagrammatic representation of various SP-A 5′UTR constructs (n = 7). Boxes labeled with A, eB, and D represent the SP-A 5′UTR exons; inv eB and ran represent the SP-A2 exon B in inverted orientation and a 30-nt random sequence, respectively. D: heterologous 5′ UTR constructs (n = 8). The boxes labeled with SP-B and SP-D represent the heterologous 5′UTRs of the human SP-B and SP-D genes, respectively. Exon B or a random sequence were cloned upstream of the heterologous 5′UTRs. eB, exon B (30 nt); ran, random sequence (30 nt); SP-B, surfactant protein B 5′UTR (14 nt), SP-D, surfactant protein D 5′UTR (43 nt).