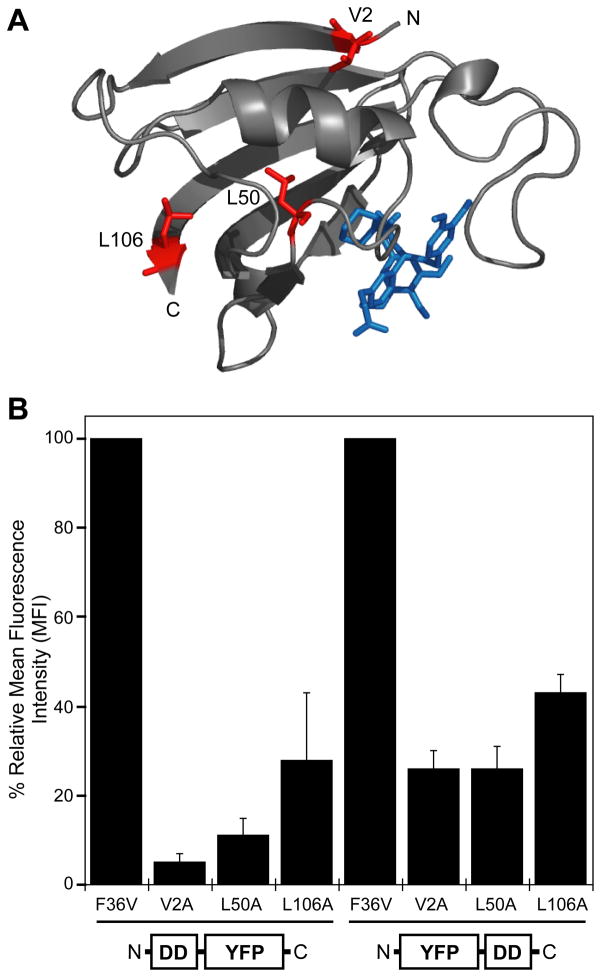
Figure 1. FKBP mutants destabilize YFP expression.
A, The structure of FKBP F36V is shown as a ribbon diagram (PyMol) complexed with the SLF* ligand (PDB ID 1BL4). Three representative mutations are highlighted in red, and the ligand is shown in blue.
B, Fluorescence of FKBP-YFP and YFP-FKBP fusions expressed in NIH3T3 cells in the absence of stabilizing ligand as determined by analytical flow cytometry. Data are presented as the average mean fluorescence intensity (MFI) ± SEM standardized to the fusions between F36V and YFP.