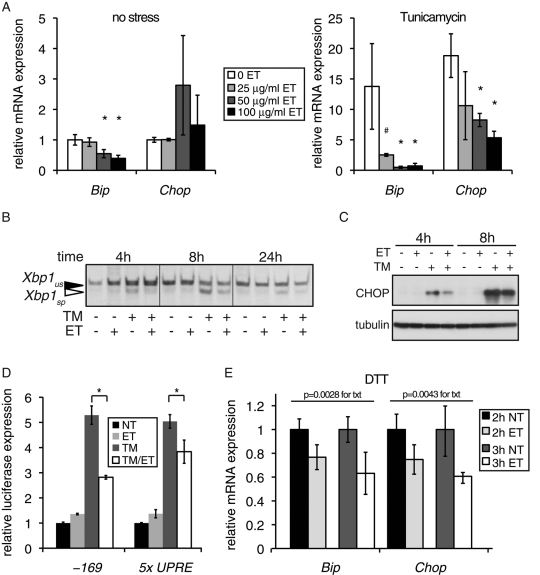
FIGURE 2:
Inhibition of FA oxidation by etomoxir attenuates UPR activation. (A) FaO hepatoma cells were treated for 6 h with the indicated doses of ET in triplicate, in the absence (left) or presence (right) of 250 ng/ml TM. Expression of Bip and Chop mRNA was quantitated by qRT-PCR. Expression is given relative to cells treated with neither chemical (note the change in scale). In this and all subsequent figures, error bars denote SDs taken from three independent treatments, unless indicated otherwise. *p < 0.05; #p < 0.1. (B) Cells were treated with 250 ng/ml TM and/or 20 μg/ml ET for the indicated times, followed by RT-PCR to detect spliced (sp) and unspliced (us) Xbp1 mRNA. (C) Cells were treated for 4 or 8 h with TM in the presence or absence of 50 μg/ml ET, and expression of CHOP was detected by immunoblot. (D) Cells were transfected with a control β-galactosidase expression plasmid and a construct containing the firefly luciferase gene controlled by either a minimal Bip promoter, encompassing the 169 proximal nucleotides and containing three ER stress response elements (–169; Luo and Lee, 2002), or by five tandem copies of a UPR element (5x UPRE; Wang et al., 2000). Cells were then treated in triplicate for 14 h with 250 ng/ml TM and/or 100 μg/ml ET, followed by quantitation of luciferase activity from cell lysates, which was normalized against β-galactosidase activity. (E) Cells were pretreated for 3 h with 50 μg/ml ET or vehicle, and then 1 mM DTT was added for a further 2 or 3 h to all samples. Relative expression of Bip and Chop mRNAs was quantitated by qRT-PCR and is given for cells pretreated with ET relative to cells that were not treated with ET. Statistical significance was determined by two-factor analysis of variance (treatment and time), with the p values for ET treatment shown.