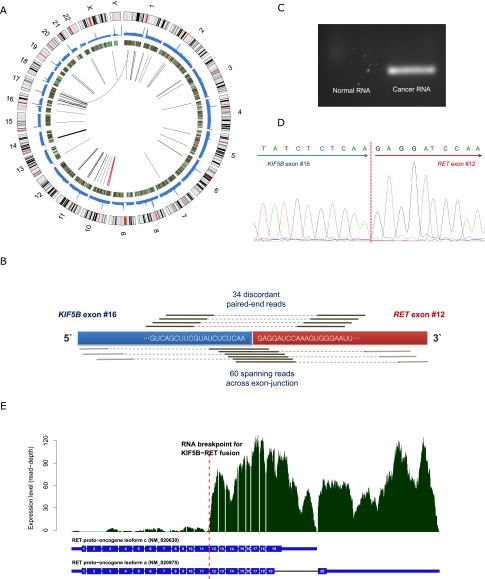
Figure 2.
Discovery of novel transforming KIF5B-RET fusion gene in lung adenocarcinoma. (A) Graphical representation of whole-genome and transcriptome sequencing data from the liver metastatic lung cancer tissue. Chromosome ideograms are shown in the outer layer. Coverage of cancer whole-genome sequencing is shown in the first middle layer. Expression level of genes is shown in the second middle layer using heatmap. Intra- and interchromosomal fusion genes are shown in the central layer. The thickness of lines shows the amount of evidence (number of spanning reads). The KIF5B-RET fusion gene is shown in red. (B) Detection of KIF5B-RET fusion gene from transcriptome sequencing. We identified 34 “discordant paired-end reads” and 60 “spanning reads” across the exon-junction. A discordant paired-end read is defined as a read whose end-sequences are aligned to each of the fusion partner genes. A spanning read is a read, one of whose end-sequences is aligned across the junction of the predicted fusion transcript. In this analysis, the fusion occurred between the 16th exon of KIF5B and 12th exon of RET. (C) Validation of KIF5B-RET fusion transcript in RNA (cDNA) from liver metastatic cancer tissue by PCR amplification and electrophoresis. The fusion gene is only detected in the liver metastatic lung cancer tissue of AK55. The negative control cDNA (normal) were extracted from the blood of a healthy Korean individual (AK1) (Kim et al. 2009). (D) Validation of the fusion gene breakpoint using Sanger sequencing in cDNA. (E) RNA expression level of each RET exon. Active expression is observed from the 12th exon, downstream from the junction of the predicted KIF5B-RET fusion gene. This suggests that the RET oncogene is expressed exclusively from the fusion gene, rather than the natural RET gene.