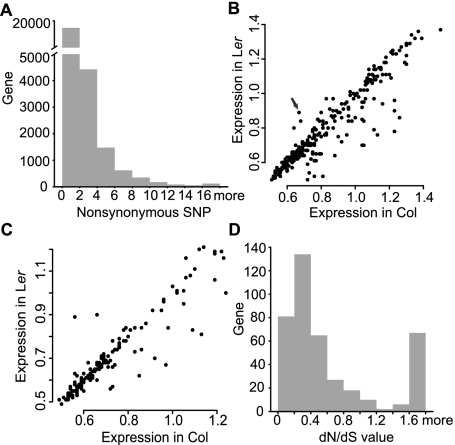
Figure 2.
Nonsynonymous SNPs and affected genes. (A) The number of nonsynonymous SNPs per gene. Although about half of the genes contain nonsynonymous SNPs, a much smaller set of genes has 10 or more nonsynonymous SNPs. (B) Expression levels in Col (x-axis) and Ler (y-axis) of genes with 10 or more nonsynonymous SNPs. The arrow points to AT5G58120, which has higher expression in Ler and encodes a disease resistance protein. (C) Expression levels in Col (x-axis) and Ler (y-axis) of genes with a Ler-premature stop codon. (D) Arabidopsis branch-specific dN/dS value of genes affected by 10 or more nonsynonymous SNPs with regard to A. lyrata. Only a few genes show neutral evolution, but most are under either positive or negative selection.