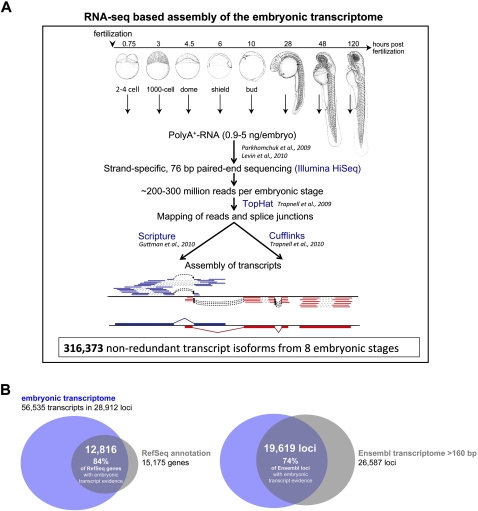
Figure 1.
Overview of the RNA-seq–based embryonic transcriptome assembly. (A) Overview of the RNA-seq–based transcript reconstruction pipeline that was employed to identify embryonically expressed transcripts in zebrafish. Stage-specific transcriptomes were reconstructed from a time-series of eight embryonic stages: two to four cell, 1000 cell, dome, shield, bud, 28 h post fertilization (hpf), 48 hpf, and 120 hpf. Stage-specific drawings of representative embryos are adapted from Kimmel et al. (1995) (with permission from Wiley © 1995). A schematic outline of the process of transcriptome reconstruction is shown at the bottom for three genes. Reads were mapped to either the + (blue) or – (red) strand using TopHat. Gaps inferred from mapping each of the two paired-end reads are indicated as dashed gray lines; dashed black arrows indicate splice-junctions inferred from a gap in mapping of a single read; and the deduced final transcript structures reconstructed by Scripture or Cufflinks are depicted at the bottom. (B) Overlap between loci from the RNA-seq–based embryonic transcriptome assembly (blue) and previously annotated genes (gray): RefSeq genes (left) and Ensembl loci >160 bp (right). The majority of known loci (84% of RefSeq loci and 74% of Ensembl loci >160 bp) are recovered in the embryonic transcriptome. Note that the number of loci in the Ensembl transcriptome is based on comparison with loci of the embryonic transcriptome (which were used as reference), which reduced the number of 27,751 Ensembl loci (>160 bp) to 26,587.