Figure A1.
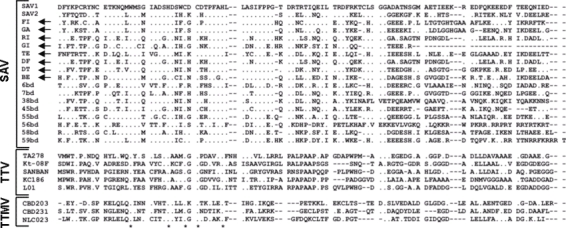
Amino acid sequences of the small anellovirus (SAV) obtained in the present study (arrows show accession nos. DQ409192 to DQ409199). GI, TE, and GA were from healthy donors; FI, RI, BE, DF, and DT were from hepatitis C patients. The sequences are aligned with all SAV present in GenBank at the time of writing and with representative torquetenovirus (TTV) and torquetenominivirus (TTMV) isolates. ClustalW alignments were manually edited and trimmed to eliminate poorly aligned positions and reduce homoplasy. As evaluated by Motif Scan and ProtParam analyses, the sequences were found to share similar properties (e.g., hydrophobicity profile, theoretical isoelectric point), irrespective of viral species. The WX7HX3CXCX5H motif highly characteristic of the open reading frame 2 of anelloviruses is indicated by asterisks.
