Figure 1. O-GlcNAc modifies the transcriptional machinery.
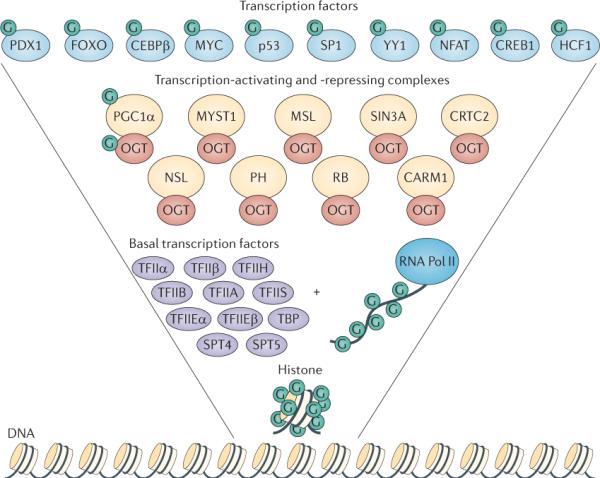
In this inverted pyramid diagram, the transcription-regulating proteins that are modified by β-D-N-acetylglucosamine (GlcNAc; G) are shown. In addition to its catalytic role, O-linked N-acetylglucosamine transferase (OGT; red ovals) interacts with numerous transcription-regulating proteins. Starting from the bottom, histones are dynamically modified by O-GlcNAc; furthermore, the carboxy-terminal domain of RNA polymerase II (RNA Pol II), as well as the basal transcription factors (purple ovals) are also modified by O-GlcNAc. Many of the transcription-activating and transcription-repressing complexes (yellow circles) that contain histone-modifying enzymes, such as histone methyltransferases, histone acetytransferases and histone deacetylases, also interact with OGT, suggesting that OGT is an integral unit in the regulation of the histone code. Finally, many transcription factors (light blue circles) are modified by O-GlcNAc. Together, the collected data from many laboratories have strongly confirmed the regulation of gene transcription by O-GlcNAcylation. CEBPβ, CCAAT/enhancer-binding protein-β; CREB1, cyclicAMP-responsive element-binding protein 1; CRTC2, CREB-regulated transcription coactivator 2; FOXO, forkhead box protein O family; HCF1, host cell factor 1; MSL, male-specific lethal; NFAT, nuclear factor of activated T cells; NSL, nonspecific lethal; PDX1, pancreas/duodenum homeobox protein 1; PGC1α: peroxisome proliferator-activated receptor-γ co-activator 1α; PH, Polyhomeotic protein; TBP, TATA-box-binding protein.
