Abstract
Certain Taraxacum species, such as Taraxacum koksaghyz and Taraxacum brevicorniculatum, produce large amounts of high-quality natural rubber in their latex, the milky cytoplasm of specialized cells known as laticifers. This high-molecular mass biopolymer consists mainly of poly(cis-1,4-isoprene) and is deposited in rubber particles by particle-bound enzymes that carry out the stereospecific condensation of isopentenyl diphosphate units. The polymer configuration suggests that the chain-elongating enzyme (rubber transferase; EC 2.5.1.20) is a cis-prenyltransferase (CPT). Here, we present a comprehensive analysis of transgenic T. brevicorniculatum plants in which the expression of three recently isolated CPTs known to be associated with rubber particles (TbCPT1 to -3) was heavily depleted by laticifer-specific RNA interference (RNAi). Analysis of the CPT-RNAi plants by nuclear magnetic resonance, size-exclusion chromatography, and gas chromatography-mass spectrometry indicated a significant reduction in rubber biosynthesis and a corresponding 50% increase in the levels of triterpenes and the main storage carbohydrate, inulin. Transmission electron microscopy revealed that the laticifers in CPT-RNAi plants contained fewer and smaller rubber particles than wild-type laticifers. We also observed lower activity of hydroxymethylglutaryl-coenzyme A reductase, the key enzyme in the mevalonate pathway, reflecting homeostatic control of the isopentenyl diphosphate pool. To our knowledge, this is the first in planta demonstration of latex-specific CPT activity in rubber biosynthesis.
Approximately 20,000 plant species produce a specialized cytoplasm known as latex, either in a laticiferous cell network or in single parenchymal cells (Bonner, 1991). The chemical composition of latex differs between species but often contains complex mixtures of isoprenoids, phenols, and alkaloids (Langenheim, 2003). Isoprenoids are structurally diverse metabolites involved in plant growth and development (sterols, brassinosteroids, ubiquinones, and dolichols) and interactions with the environment (sesquiterpenoid phytoalexins and monoterpenes). Natural rubber is an economically valuable isoprenoid found in the latex cells of approximately 2,500 plant species (Mooibroek and Cornish, 2000).
The high-performance natural rubber favored by industry comprises more than 10,000 cis-1,4-linked isoprene units producing a biopolymer with a molecular mass exceeding 500,000 D. This is synthesized in plants such as Hevea brasiliensis, Parthenium argentatum, Ficus species, and especially in the roots of Taraxacum species (for review, see van Beilen and Poirier, 2007; Munt et al., 2012). Large quantities of isopentenyl diphosphate (IPP) are required for rubber biosynthesis, and this is thought to be derived from the mevalonate (MVA) pathway, as indicated by radioactive labeling assays using latex extracts from H. brasiliensis (Bandurski and Teas, 1957; Hepper and Audley, 1969). Enzymes in the plastid-localized methylerythritol phosphate (MEP) pathway are also expressed in H. brasiliensis latex (Seetang-Nun et al., 2008). However, there was no significant incorporation of [1-13C]1-deoxyxylulose into cis-polyisoprene in H. brasiliensis seedlings, even though this compound is closely related to the early MEP pathway intermediate 1-deoxyxylulose 5-phosphate, whereas large amounts of labeled carotenoids were detected in the same experiment, suggesting that the H. brasiliensis MEP pathway contributes to diterpenoid and carotenoid biosynthesis but not rubber biosynthesis (Sando et al., 2008).
Although there are significant differences between the various rubber-producing plant species, the basic mechanisms of natural rubber biosynthesis are similar, involving comparable enzymes (Cornish, 2001; Bushman et al., 2006; Schmidt et al., 2010a, 2010b) that are compartmentalized in organelle-like rubber particles in the latex (Cornish et al., 1999; Wood and Cornish, 2000; Cornish, 2001). When Ficus species rubber particles are incubated in the presence of [1-14C]IPP and an allylic diphosphate, the isoprenoid chains are elongated by a rubber transferase (EC 2.5.1.20) with cis-prenyltransferase (CPT) activity (Siler and Cornish, 1993; Kang et al., 2000). CPTs are further divided into those producing short, medium, and long isoprene chains, and they differ from transprenyltransferases (TPTs) in the configuration of the double bond formed during IPP condensation and the absence of sequence or structural similarity despite sharing the same substrate (Takahashi and Koyama, 2006; Lu et al., 2010; Punetha et al., 2010).
Many TPTs have already been identified, and the Arabidopsis (Arabidopsis thaliana) farnesyl diphosphate synthase, which catalyzes the formation of E,E-farnesyl diphosphate (FPP; Lange and Ghassemian, 2003; Bouvier et al., 2005), is particularly well characterized (Cunillera et al., 1996, 1997; Manzano et al., 2006; Closa et al., 2010). In contrast, relatively few CPTs have been isolated and characterized, including CPTs from Arabidopsis (Cunillera et al., 2000; Oh et al., 2000; Zhang et al., 2008; for review, see Surmacz and Swiezewska, 2011) and two CPTs from H. brasiliensis, the Hevea rubber transferases (HRT1 and HRT2; Asawatreratanakul et al., 2003). HRT1 and HRT2 are predominantly expressed in the latex, and recombinant HRT2 (but not HRT1) can produce high-molecular mass natural rubber in the presence of washed bottom-fraction particles from Hevea latex, IPP, allylic diphosphates such as FPP, and a divalent cation such as Mg2+ or Mn2+ as a cofactor (Asawatreratanakul et al., 2003). However, there has been no direct confirmation of the role of CPT in rubber biosynthesis in planta thus far.
Enzymatically active rubber particles were recently isolated from Taraxacum brevicorniculatum (previously described as Taraxacum koksaghyz; Schmidt et al., 2010b), and their CPT activity was confirmed by the incorporation of labeled IPP. Three highly conserved CPTs named TbCPT1 to -3 were identified, and these were found to be expressed predominantly in laticifers and associated with rubber particles (Schmidt et al., 2010a, 2010b). All three enzymes were also able to produce long-chain poly(cis-1,4-isoprene) in yeast.
Here, we demonstrate the crucial role of TbCPT1 to -3 in T. brevicorniculatum by using RNA interference (RNAi) in transgenic plants to silence the corresponding genes. This inhibited the synthesis of high-molecular mass natural rubber and the formation of regular rubber particles and resulted in elevated levels of triterpenes in the latex (for triterpenes in Taraxacum species, see Furuno et al., 1993; Akashi et al., 1994) and of the major storage carbohydrate inulin in the roots (for inulin in Taraxacum species, see Van den Ende et al., 2000; Schütz et al., 2006). The redirection of carbon flux in CPT-RNAi plants to increase the levels of inulin and nonrubber isoprenoids is discussed in the context of our current understanding of the regulatory mechanisms controlling the isoprenoid pathway.
RESULTS
Latex-Specific Suppression of TbCPT1 to -3 Expression
Three highly conserved CPTs were recently isolated from T. brevicorniculatum (TbCPT1 to -3; Schmidt et al., 2010a). To gain more insight into their precise activities, we investigated the role of TbCPT1 to -3 in latex by generating transgenic T. brevicorniculatum plants in which all three CPT genes were silenced by RNAi. A TbCPT-RNAi construct was assembled containing a hairpin 491-bp TbCPT cDNA fragment (nucleotides 118–608, encompassing four of the five motifs that distinguish CPTs from TPTs; Kharel and Koyama, 2003) under the control of the laticifer-specific Taraxacum officinale Polyphenoloxidase1 (PPO1) promoter (Wahler et al., 2009; Fig. 1A). This strategy was chosen to prevent the silencing of nonlatex CPTs, which may be necessary for housekeeping functions in other cell types. T. brevicorniculatum plants were transformed using the Agrobacterium tumefaciens leaf disc method, leading to the recovery of 12 independent transgenic lines.
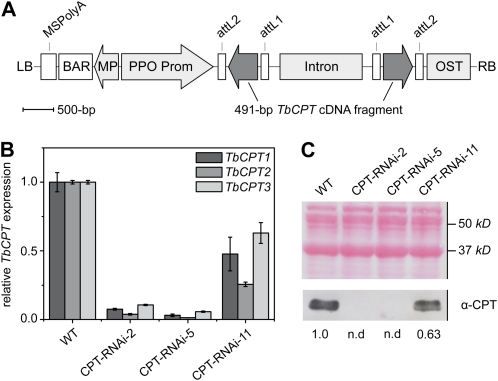
Figure 1.
RNA silencing of TbCPT1 to -3 in transgenic T. brevicorniculatum CPT-RNAi lines. A, Schematic representation of the CPT-RNAi vector. The 491-bp TbCPT cDNA fragment was used as a trigger for RNAi under the control of the T. officinale latex-specific PPO promoter (PPO Prom). attL1/2, attL cassette frame; BAR, basta resistance gene; Intron, chalcone synthase intron from P. hybrida; LB, left border; MP, mannopine synthase promoter; MSPolyA, mannopine synthase poly(A) signal; OST, octopine synthase poly(A) signal; RB, right border. B, Quantitative RT-PCR analysis of TbCPT1 to -3 mRNA expression in the latex from roots of transgenic CPT-RNAi lines and in a wild-type (WT) control. The expression levels were normalized to TbGAPDH and set as 1 for the wild type. C, Protein analysis of transgenic CPT-RNAi lines and a wild-type control. Latex samples were used for protein separation by SDS-PAGE and subsequent western-blot analysis. The top image shows Ponceau S staining of the nitrocellulose membrane after protein transfer. This membrane was used for a western blot using a TbCPT-specific α-CPT antibody (bottom image). Numbers below the blot indicate signal intensities relative to the wild-type value. n.d., No signal detectable above background. [See online article for color version of this figure.]
We confirmed the presence of the transgene in each line by PCR and analyzed the expression of each TbCPT gene by quantitative reverse transcription (RT)-PCR (Fig. 1B; Supplemental Fig. S1) using specific primers for TbCPT1 to -3 to amplify total RNA isolated from the latex of roots and petioles. The efficiency of CPT knockdown varied among the different lines (eight strong, two intermediate, and two unaffected). Therefore, we were able to select three lines with different residual TbCPT expression levels for further analysis: CPT-RNAi-2 and CPT-RNAi-5, which demonstrated almost complete silencing, and CPT-RNAi-11, which demonstrated partial silencing. The transgenic lines were morphologically similar to wild-type plants. In all three lines, the relative residual TbCPT1 to -3 expression levels followed a similar profile, with TbCPT3 expressed at the highest level followed by TbCPT1 and then TbCPT2.
We also determined the efficiency of TbCPT1 to -3 knockdown at the protein level by western-blot analysis (Fig. 1C). Total latex proteins were separated by SDS-PAGE and transferred to a nitrocellulose membrane for immunodetection with a polyclonal anti-TbCPT antibody recognizing all three proteins, revealing TbCPT1 and TbCPT2 bands with a molar mass of 34 kD and a 32-kD TbCPT3 band (Schmidt et al., 2010b). Whereas TbCPT1 to -3 protein levels were only partially reduced in CPT-RNAi-11 plants, none of the proteins were detected in latex extracts from transgenic lines CPT-RNAi-2 and CPT-RNAi-5.
To understand the relationship between the amount of TbCPT protein in the latex and the overall prenyltransferase activity, we carried out in vitro [1-14C]IPP incorporation assays with crude latex extracts from each of the transgenic lines (and wild-type controls) as described by Schmidt et al. (2010b). Short- and medium-chain polyprenol diphosphates were removed by extraction with n-butanol as described by Asawatreratanakul et al. (2003), and then long-chain products were extracted with a mixture of toluene and hexane (Table I). We found that in vitro long-chain prenyltransferase activity was detected only following the addition of whole latex extracts from a wild-type plant with a [1-14C]IPP conversion rate of 0.62 pmol min−1 mg−1 or line CPT-RNAi-11 with a [1-14C]IPP conversion rate of 0.5 pmol min−1 mg−1 (reduced by approximately 19%), whereas no long-chain radiolabeled products were synthesized in lines CPT-RNAi-2 and CPT-RNAi-5.
Table I. Influence of CPT-RNAi on [1-14C]IPP incorporation into long-chain polyisoprenes in vitro.
The [1-14C]IPP incorporation assay was performed using 50 μg of whole latex protein from CPT-RNAi and wild-type plants. The negative control contained only E,E-FPP and [1-14C]IPP as substrates. Long-chain products were extracted with a mixture of toluene and hexane after the extraction of short- and medium-chain polyprenol diphosphates with n-butanol, and the incorporation of labeled IPP was determined using a scintillation counter. The values represent three independent experiments ± sd.
| Plant | Butanol Extract | Toluene/Hexane Extract |
| Bq | ||
| Negative control | 144.4 | 3.5 |
| Wild type | 287.3 ± 28.5 | 46.3 ± 3.9 |
| CPT-RNAi-2 | 278.3 ± 7.7 | 3.0 ± 0.4 |
| CPT-RNAi-5 | 246.6 ± 9.5 | 2.6 ± 0.7 |
| CPT-RNAi-11 | 279.6 ± 7.8 | 38.5 ± 1.3 |
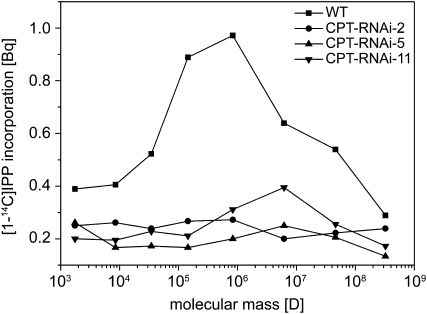
To determine the molecular mass of the long-chain products synthesized in wild-type and CPT-RNAi-11 plants, we hydrolyzed the products after the assay reaction and fractionated total hexane extracts after further purification steps by size-exclusion chromatography (SEC; Fig. 2). This revealed in a qualitative manner that long-chain polyisoprenes with a molecular mass between 105 and 107 D were produced in wild-type plants, whereas line CPT-RNAi-11 contained lower levels of long-chain polyisoprenes (particularly those with a molecular mass of approximately 107 D). The total hexane extracts for lines CPT-RNAi-2 and CPT-RNAi-5 contained no labeled long-chain products at all, which is consistent with the two-step differential solvent extraction of radiolabeled products as described above.
Figure 2.
Molecular mass distribution of the labeled assay products from the in vitro [1-14C]IPP incorporation assay. The products were hydrolyzed after the assay, extracted with hexane, and fractionated by SEC, and radioactivity in the resulting fractions was determined using a scintillation counter. WT, Wild type.
TbCPT1 to -3 Knockdown Suppresses Rubber Biosynthesis in Planta
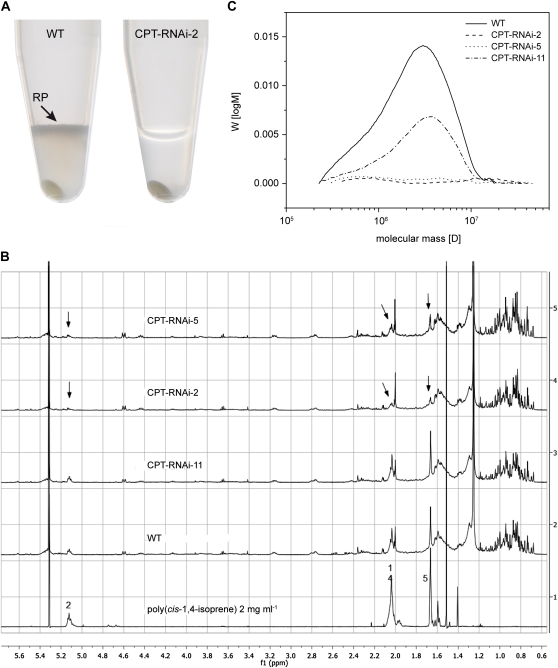
We next analyzed the quantity and characteristics of T. brevicorniculatum rubber particles to gain more insight into the roles of TbCPT1 to -3 in planta. We harvested the latex from wild-type and transgenic T. brevicorniculatum plants in rubber extraction buffer and then centrifuged the solution to yield a pellet phase, the aqueous cytoplasm (C-serum), and an upper rubber phase containing rubber particles (Schmidt et al., 2010b). Whereas all three phases were present in wild-type latex (Fig. 3A), there was no rubber phase in the latex from lines CPT-RNAi-2 and CPT-RNAi-5 (Fig. 3A shows CPT-RNAi-2 as an example), clearly demonstrating the crucial role of TbCPT1 to -3 in the synthesis of natural rubber in planta.
Figure 3.
Influence of CPT silencing on rubber content and rubber composition. A, Reaction tubes (1.5 mL) containing latex extracts in rubber extraction buffer after centrifugation. The upper rubber phase (RP) was not present in latex from lines CPT-RNAi-2 and CPT-RNAi-5 (CPT-RNAi-2 is shown as an example). B, 1H-NMR spectra of root extracts from wild-type (WT) and CPT-RNAi T. brevicorniculatum plants using hexane as the solvent. The NMR spectra were scaled to the solvent signal at 5.3 ppm. The 1H-NMR spectrum of a poly(cis-1,4-isoprene) reference sample is also shown for comparison. The arrows in the top traces indicate residual signals of the cis-configured polymer. The numbers relate to positions of poly(cis-1,4-isoprene) given in Supplemental Figure S2. C, SEC analysis of latex extracts from wild-type and CPT-RNAi lines. Whole latex was used for the hexane extraction of hydrophobic compounds. Molecular mass was determined using poly(cis-1,4-isoprene) standards. [See online article for color version of this figure.]
We next determined the quantity of poly(cis-1,4-isoprene) in wild-type and CPT-RNAi root extracts by 1H-NMR spectroscopy (Schmidt et al., 2010b). Strong poly(cis-1,4-isoprene) signals were detected in wild-type extracts, but lines CPT-RNAi-2 and CPT-RNAi-5 produced much weaker signals (Fig. 3B). The amount of poly(cis-1,4-isoprene) per gram dry weight of root tissue was determined by comparing the peak intensities of the C5 methyl protons (Schmidt et al., 2010b; Supplemental Fig. S2A) with the corresponding signals for a poly(cis-1,4-isoprene) reference. This indicated that the quantity of poly(cis-1,4-isoprene) in extracts from lines CPT-RNAi-2, CPT-RNAi-5, and CPT-RNAi-11 was 25%, 38%, and 84% relative to wild-type extracts, respectively (Table II). We also determined the dry rubber content by gravimetric analysis. Freeze-dried root material was extracted with toluene to dissolve long-chain poly(cis-1,4-isoprene), and this was precipitated with methanol. The mass of each precipitate revealed that lines CPT-RNAi-2 and CPT-RNAi-5 contained very limited amounts of long-chain poly(cis-1,4-isoprene): 7% and 4% of the wild-type amount, respectively (Table II). The combined NMR and gravimetric analysis data indicate that lines CPT-RNAi-2 and CPT-RNAi-5 are strongly inhibited in their capacity to produce long-chain poly(cis-1,4-isoprene).
Table II. Quantitative determination of poly(cis-1,4-isoprene) by gravimetric analysis and 1H-NMR spectroscopy.
Quantification was performed using freeze-dried root material from CPT-RNAi and wild-type plants. Dry rubber content was measured following toluene extraction, methanol precipitation of long-chain poly(cis-1,4-isoprene), and gravimetric analysis of the precipitate. The total values represent the analysis of three samples per line. For 1H-NMR spectroscopy, the quantity of poly(cis-1,4-isoprene) g−1 dry weight of root tissue in each of the lines was determined by comparing the peak intensities of the methyl protons with the corresponding signals of a poly(cis-1,4-isoprene) reference.
| Plant | Gravimetric Analysis | NMR | ||
| mg g−1 dry wt | % | mg g−1 dry wt | % | |
| Wild type | 1.79 | 100 | 3.10 | 100 |
| CPT-RNAi-2 | 0.12 | 7 | 0.75 | 25 |
| CPT-RNAi-5 | 0.07 | 4 | 1.18 | 38 |
| CPT-RNAi-11 | 1.49 | 83 | 2.59 | 84 |
Direct evidence of poly(cis-1,4-isoprene) chain length was provided by additional SEC analysis of latex extracts to determine their weight average molecular mass (Mw) and number average molecular mass (Mn). As shown in Figure 3C, a large quantity of long-chain poly(cis-1,4-isoprene) was detected in the wild-type extract, with a molecular mass between 106 and 107 D, a Mw of 4.73 × 106 D, and a Mn of 3.32 × 106 D. In agreement with the NMR and gravimetric analysis data, the quantity of poly(cis-1,4-isoprene) was reduced in line CPT-RNAi-11, but the molecular mass distribution values were similar to the wild type (Mw = 5.44 × 106 D; Mn = 3.79 × 106 D), indicating that polymer length and polydispersity were unaffected by the reduced TbCPT enzyme activity. In contrast, no high-molecular mass rubber chains were detected in lines CPT-RNAi-2 or CPT-RNAi-5. These data indicate that the poly(cis-1,4-isoprene) signal in the NMR spectra that was not detected either by gravimetric analysis or SEC represented short-chain poly(cis-1,4-isoprene) molecules and that there is almost no rubber produced in the transgenic lines.
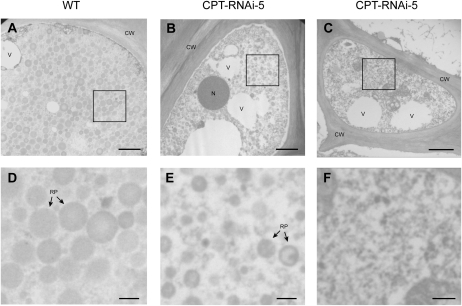
Next, we investigated whether the absence of long-chain poly(cis-1,4-isoprene) in the CPT-RNAi lines coincided with the loss of rubber particles in the latex, which would explain the absence of the rubber phase in centrifuged latex extracts from lines CPT-RNAi-2 and CPT-RNAi-5 (Fig. 3A). We studied the morphology of rubber particles within the root laticifers of each line by transmission electron microscopy (TEM). This revealed abundant and closely packed rubber particles in wild-type plants (Fig. 4, A and D) but fewer particles in the transgenic lines (Fig. 4, B and E), none in some cases (Fig. 4, C and F), and these particles tended to be smaller than their wild-type counterparts. The less abundant rubber particles in the transgenic lines, therefore, behaved differently from those from the wild-type plants during separation and did not form a typical rubber phase.
Figure 4.
TEM images of a root laticifer from wild-type (WT; A) and transgenic line CPT-RNAi-5 (B and C) plants. Bottom images (D–F) show higher magnifications of the details indicated by the squares above. CW, Cell wall; N, nucleus; RP, rubber particles; V, vacuole. Bars = 1 μm in A to C and 0.2 μm in D to E. [See online article for color version of this figure.]
Rubber Biosynthesis Is Linked to Triterpene and Inulin Content
Natural rubber in T. brevicorniculatum latex is derived from IPP, so it is likely that the inhibition of rubber biosynthesis enlarges the IPP pool, providing more precursors for other isoprenoid pathways. We investigated whether the increase in IPP levels had any impact on these other pathways and whether homeostatic controls came into effect.
Taraxacum species latex contains abundant quantities of pentacyclic triterpenes, including some molecules found in many other plants, such as lupeol and amyrins, and other molecules that are largely restricted to the genus Taraxacum, such as taraxasterol (Furuno et al., 1993; Akashi et al., 1994; Supplemental Fig. S2B). To determine the differences in triterpene levels between transgenic and wild-type plants, we measured the levels of these different triterpenes in hexane extracts of wild-type and transgenic roots with a gas chromatograph equipped with a flame ionization detector (GC/FID), and we found that the total triterpene levels were significantly higher in CPT-RNAi roots compared with wild-type roots (i.e. 80% higher in CPT-RNAi-2, 75% higher in CPT-RNAi-5, and 18% higher in CPT-RNAi-11; Table III). In addition to the pentacyclic triterpenes, the levels of sterols such as cholesterol, sitosterol, and stigmasterol had also increased.
Table III. Influence of CPT-RNAi on the triterpene content of T. brevicorniculatum roots.
Triterpene levels in hexane root extracts were quantified by GC/FID, and structures were confirmed by GC-MS. The total triterpene values represent the analysis of three samples per line with low variation (sd less than 1%). The sterol and pentacyclic triterpene values are from one representative of the three samples per line.
| Constituent | Wild Type | CPT-RNAi-2 | CPT-RNAi-5 | CPT-RNAi-11 |
| mg g−1 dry wt | ||||
| Total C30 derivatives | 2.61 | 4.69 | 4.57 | 3.09 |
| Sterols | ||||
| 24-Methyl cholesterol | 0.05 | 0.10 | 0.10 | 0.09 |
| Sitosterol | 0.30 | 0.39 | 0.39 | 0.33 |
| Stigmasterol | 0.12 | 0.21 | 0.19 | 0.15 |
| Pentacyclic triterpenes | ||||
| Lupeol | 0.14 | 0.25 | 0.24 | 0.16 |
| β-Amyrin | 0.26 | 0.51 | 0.52 | 0.32 |
| α-Amyrin | 0.23 | 0.45 | 0.44 | 0.30 |
| Taraxerol | 0.28 | 0.34 | 0.24 | 0.25 |
| Ψ-Taraxasterol | 0.32 | 0.60 | 0.61 | 0.39 |
| Taraxasterol | 0.53 | 1.03 | 1.04 | 0.65 |
| Unidentified triterpenes | 0.38 | 0.81 | 0.80 | 0.45 |
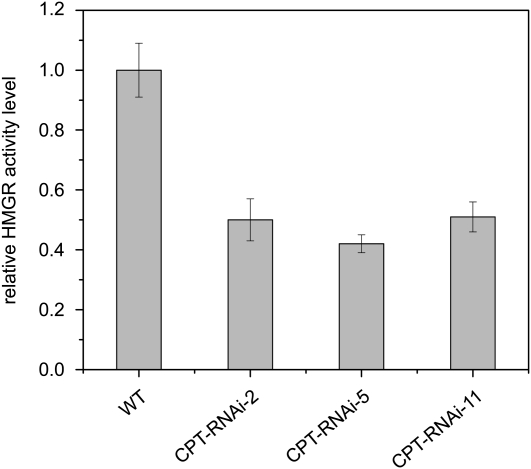
Despite the above data, the higher triterpene content of the transgenic lines cannot on its own explain what becomes of the excess IPP in plants when rubber synthesis is completely inhibited. Therefore, we investigated additional regulatory models. The MVA pathway is thought to be the main source of IPP in rubber-producing plants (Bandurski and Teas, 1957; Hepper and Audley, 1969; Sando et al., 2008), so we studied the expression of hydroxymethylglutaryl-coenzyme A reductase (HMGR), the rate-limiting enzyme of the MVA pathway (Chappell et al., 1995). This enzyme is known to be feedback regulated by isoprenoid compounds (for review, see Stermer et al., 1994). We measured the HMGR activity in latex from wild-type and transgenic plants using [3-14C]HMG-CoA as the substrate and found a significant reduction in the activity of this enzyme in all transgenic lines (Fig. 5), presumably resulting from feedback inhibition either by IPP itself or by mevalonate- or squalene-derived end products.
Figure 5.
HMGR activity in the latex of wild-type (WT) and CPT-RNAi plants as determined by [3-14C]HMG-CoA incorporation assay. HMGR activity was calculated from the amount of labeled HMG-CoA incorporated into mevalonolactone mg−1 protein h−1, and the value of the wild type was set to 1. The values presented are from two independent experiments including three technical replicates ± sd. HMGR activity was significantly lower in the CPT-RNAi lines as calculated using Student’s t test (P < 0.01).
Having established that the inhibition of rubber biosynthesis reduces flux through the MVA pathway, we next addressed the fate of the earlier precursors that feed into this pathway, such as acetyl-CoA and acetoacetyl-CoA. Because the main carbon storage polymer in dandelion (Taraxacum spp.) roots is inulin (Van den Ende et al., 2000; Schütz et al., 2006), we investigated whether the carbon units originally destined for the MVA pathway might be redirected to this end product. Therefore, we determined the total amount of inulin in crude aqueous extracts of wild-type and transgenic root material by 1H-NMR using established NMR-based metabolomics protocols that allow the quantitation of inulin without prior separation or chemical processing. The 1H-NMR spectra for each extract showed predominant signals for inulin (Cérantola et al., 2004). The absolute concentration of inulin was then determined on the basis of the well-resolved inulin signal at 4.04 ppm (corresponding to position H4 of the fructosyl group [Cérantola et al., 2004; Supplemental Fig. S2C]) relative to an internal reference. We found that wild-type roots contained 554 ± 46 mg inulin g−1 dry tissue, line CPT-RNAi-11 had similar levels (566 ± 83 mg g−1), and the transgenic lines, with almost complete suppression of rubber biosynthesis, had significantly higher levels (675 ± 47 mg g−1 for line CPT-RNAi-2 and 642 ± 117 mg g−1 for line CPT-RNAi-5; Table IV; Supplemental Table S1).
Table IV. Relative amounts of inulin (normalized to wild-type plants set to 100) in the roots of T. brevicorniculatum as determined by 1H-NMR and spectrophotometry after acid hydrolysis.
The t values were calculated using the averaged values (n = 15). Statistical significance with a 99% confidence level was achieved for t > 2.8. n indicates the number of independent measurements.
| Plant | Photometry (n = 6) | NMR (n = 9) | Averaged (n = 15) | t Value (n = 15) |
| Wild type | 100 | 100 | 100 | |
| CPT-RNAi-2 | 113 | 122 | 117 | 6.5 |
| CPT-RNAi-5 | 108 | 116 | 112 | 3.4 |
| CPT-RNAi-11 | 94 | 102 | 98 | 0.7 |
We used an independent method based on the enzymatic hydrolysis of root extracts and the photometric quantitation of Fru and Glc to confirm the inulin levels in the four lines. The absolute values obtained by this method were considerably lower than those determined by 1H-NMR (Supplemental Table S1), but this may reflect the partial degradation of Fru under the experimental conditions used for inulin hydrolysis resulting in the underestimation of inulin levels (Szambelan and Nowak, 2006; Nasab et al., 2009; Nguyen et al., 2009). Therefore, we normalized the amounts of inulin to relative values, setting the wild-type level arbitrarily to 100 (Table IV). Both the 1H-NMR and in vitro hydrolysis data showed that lines CPT-RNAi-2 and CPT-RNAi-5 contained 10% to 20% more inulin than wild-type plants, whereas the amount of inulin in CPT-RNAi-11 plants remained similar to that in the wild type. Student’s t test confirmed 99% confidence in the data for lines CPT-RNAi-2 and CPT-RNAi-5.
DISCUSSION
Natural rubber is a valuable polymer with useful properties, such as high elasticity, resilience, and thermal stability, which cannot be matched by synthetic alternatives. Most of the world’s natural rubber is currently sourced from the Pará rubber tree (H. brasiliensis), but other species such as the desert shrub P. argentatum and dandelions are potential alternatives. A major advantage of dandelions is their suitability for genetic manipulation, which can also provide insights into the mechanisms of rubber biosynthesis. Therefore, we chose T. brevicorniculatum as a model to demonstrate, to our knowledge for the first time, that CPTs are required for the final step in rubber biosynthesis [i.e. the elongation of poly(cis-1,4-isoprene) chains by the addition of IPP precursors].
We recently isolated three T. brevicorniculatum CPT genes that we named TbCPT1 to -3 (Schmidt et al., 2010a). We showed that the corresponding enzymes are compartmentalized on rubber particles within the latex and that recombinant versions catalyze the formation of long-chain poly(cis-1,4-isoprene) in yeast (Schmidt et al., 2010b).
Although there is plenty of evidence supporting the role of TbCPT1 to -3 in rubber biosynthesis, this has not yet been demonstrated directly in planta. Therefore, we used RNAi to silence the three corresponding genes and investigated the impact on rubber biosynthesis. T. brevicorniculatum plants provide an ideal model for such studies because they produce high-quality rubber, have a short life cycle, and can be genetically modified using a simple and reliable protocol (Wahler et al., 2009).
The almost complete silencing of TbCPT1 to -3 in lines CPT-RNAi-2 and CPT-RNAi-5 was confirmed at the mRNA and protein levels (Fig. 1, B and C), and this reduced the overall poly(cis-1,4-isoprene) content by up to 75% and led to the almost complete absence of long-chain poly(cis-1,4-isoprene) molecules, as shown by SEC (Fig. 3C). The partial silencing of TbCPT1 to -3 in line CPT-RNAi-11 showed that the quantity of long-chain poly(cis-1,4-isoprene) correlated directly with the availability of these enzymes. Line CPT-RNAi-11 contained approximately 40% of the wild-type level of TbCPT1 to -3 (Fig. 1B), resulting in a weaker CPT signal on western blots (Fig. 1C) and weaker in vitro long-chain prenyltransferase activity in the [1-14C]IPP incorporation assay (Table I). The lower CPT activity in line CPT-RNAi-11 reduced the amount of high-molecular mass rubber (Fig. 3C), but physicochemical properties such as polymer length and polydispersity were unaffected. The almost complete silencing in lines CPT-RNAi-2 and CPT-RNAi-5 resulted in the absence of a western-blot signal (Fig. 1C) and a complete loss of in vitro long-chain prenyltransferase activity (Table I). Taken together, the CPT-silencing data indicate that rubber biosynthesis in T. brevicorniculatum latex is fully dependent on CPT activity.
Asawatreratanakul et al. (2003) showed that additional latex proteins or cofactors are required for the production of high-molecular mass rubber in H. brasiliensis. They carried out in vitro [1-14C]IPP incorporation assays with the recombinant CPTs HRT1 and HRT2 and found that purified HRT2 (but not HRT1) produces high-molecular mass (105 and 106 D) polyisoprenes only in the presence of washed bottom-fraction particles from H. brasiliensis latex. Although the nature of these additional factors remains elusive, our data confirm that CPTs are an absolute requirement for the synthesis of long polyisoprene chains.
Another striking characteristic of CPT-depleted latex was the complete loss of the rubber phase (Fig. 3A), reflecting the reduced number of rubber particles (Fig. 4). Ultrastructural analysis of rubber particles from different species has revealed an organelle-like structure comprising a homogeneous hydrophobic rubber core surrounded by a monolayer membrane that contains lipids, proteins, and additional minor compounds, which differ between species (Cornish et al., 1999; Wood and Cornish, 2000; Cornish, 2001). Rubber particles are structurally similar to the lipid bodies found in all eukaryotic cells, which comprise a hydrophobic core of neutral lipids surrounded by a phospholipid monolayer containing various proteins (for review, see Martin and Parton, 2006). These bodies are thought to function as lipid storage organelles, although they may also participate directly in lipid synthesis (Sato et al., 2001). Lipid bodies are thought to form by budding off from discrete regions of the endoplasmic reticulum (ER) membrane (Huang, 1992). In this model, newly formed neutral lipids that are unable to integrate into a phospholipid bilayer accumulate between the two phospholipid layers of the ER membrane as microdroplets, which bud off once they reach a certain size to form a nascent lipid body. The major phospholipid in the monolayer of a lipid body is phosphatidylcholine (Huang, 1992), and this molecule is also abundant in the rubber particle monolayers of different rubber-producing plants (Cornish et al., 1999). These structural similarities suggest that lipid bodies and rubber particles form in the same manner, and we propose that the inhibition of rubber synthesis leads to fewer initiation events on the ER membrane and therefore to the lower number of rubber particles in the CPT-RNAi plants. The budding hypothesis for the biogenesis of rubber particles remains to be confirmed.
The inhibition of rubber biosynthesis leads to the accumulation of precursors; therefore, we carried out experiments to determine how the carbon flux is redirected. We showed that in addition to the synthesis of pentacyclic triterpenes, which are highly abundant in the latex of Taraxacum species (Furuno et al., 1993; Akashi et al., 1994), the sterol pathway is stimulated by the inhibition of CPT activity in the laticifers, resulting in an increase of up to 80% in the total triterpene content in the roots. This suggested that root tissues of the CPT-RNAi lines (and not only laticiferous cells) developed an integrated metabolic response to the enlarged IPP pool in poly(cis-1,4-isoprene)-depleted cells. The formation of esterified sterols is one regulatory process that maintains sterol homeostasis in the plant cell, and plants can store large amounts of sterols as sterol esters in lipid bodies (Schaller et al., 1995; Bouvier-Navé et al., 2010). Another mechanism is the feedback inhibition of HMGR, the key rate-limiting enzyme of the MVA pathway (Stermer et al., 1994). Three HMGRs were recently identified in T. brevicorniculatum (TbHMGR1 to -3), and expression analysis indicated an important role for TbHMGR1 in the provision of precursors for rubber biosynthesis in the latex (van Deenen et al., 2011). We showed that HMGR enzyme activity was inhibited in the latex of the CPT-RNAi plants, presumably either by the accumulation of sterols or by IPP itself. The feedback regulation of HMGR has been reported previously. For example, positive feedback was reported in tobacco (Nicotiana tabacum) Bright Yellow 2 cells treated with specific inhibitors of squalene synthase and squalene epoxidase (the first two steps in the sterol synthesis pathway), increasing the level of HMGR activity (Wentzinger et al., 2002).
The inhibition of HMGR reduces the synthesis of IPP but in turn leads to the accumulation of upstream precursors such as acetyl-CoA, which is used to generate energy through the citric acid cycle. Energy is stored when it is not required, and the major storage carbohydrate in dandelion roots is inulin (Van den Ende et al., 2000; Schütz et al., 2006). Therefore, we investigated the inulin content in transgenic and wild-type plants and found that, in transgenic plants with almost complete inhibition of rubber biosynthesis, the inulin content was increased by up to 20%. The ability of dandelion plants to absorb the excess IPP resulting from the lack of rubber biosynthesis, and to use feedback regulation to direct the flux efficiently into another storage product, shows remarkable adaptability and explains why the transgenic plants showed no developmental abnormalities. Interestingly, the inulin content of transgenic line CPT-RNAi-11 and wild-type plants was similar, despite the much more significant impact on sterol levels and HMGR activity. This suggests that the redirection of excess IPP to the sterol biosynthesis pathway is the first and immediate response to the suppression of rubber synthesis, and only when this relief pathway is also saturated (in plants with nearly complete CPT silencing) does the buildup of precursors for the MVA pathway affect upstream flux and redirect carbon to the storage product inulin. Whether the redirection of carbon flux was a direct consequence of rubber depletion and/or the increase in sterol levels or the consequence of off-target suppression or pleiotropic effects has yet to be determined.
We have presented to our knowledge the first in planta demonstration of the pivotal role of CPTs in the production of natural rubber, and this provides a clearer insight into the regulatory mechanisms governing the relationship between rubber biosynthesis and connected metabolic pathways.
MATERIALS AND METHODS
Plant Material and Cultivation Conditions
Taraxacum brevicorniculatum plants were obtained from the Karlsruhe Botanical Gardens and were cultivated at 18°C with a 16-h photoperiod (20 klux) in controlled growth chambers or in the greenhouse. Plants were cultivated in a prefertilized 1:1 mixture of standard soil (ED73 Einheitserde) and garden mold (Münster Botanical Garden) and fertilized every 4 weeks with a commercial fertilizer according to the manufacturer’s recommendations (Hakaphos Plus; Compo).
Assembling the CPT-RNAi Construct
The Taraxacum officinale PPO1 promoter (Wahler et al., 2009) was inserted into vector pLab12.1 (linearized with XmaI and XhoI) to create pLab12.4. The pLab12.1 vector was originally constructed by digesting pFGC5941GW (Wahler et al., 2009) with the enzymes EcoRI and PmeI and inserting the linker sequence 5′-AATTAAGCTTCCCGGGTACCTCGCGACTCGAGCCATGGTCTAGAGGATCCGAATTCGTTTAAACGAATTCGGATCCTCTAGACCATGGCTCGAGTCGCGAGGTACCGGAAGCTT-3′. A cassette from pLab12.4 containing the PPO1 promoter was inserted into the KpnI and XhoI sites of the pFGC5941 RNAi cassette (http://www.chromDB.org), which contains the Petunia hybrida chalcone synthase intron, resulting in the RNAi vector PFGC-PPOprom. A 491-bp CPT PCR fragment was amplified using the RNAi-dicer optimized primers RNAi-for (5′-AAAGGTACCGTGTTTCGTGTCATTGCTTC-3′) and RNAi-rev (5′-AAACTCGAGGCAACACATACGAGGAGATACG-3′) and was inserted into the KpnI and XhoI sites of the Gateway vector pENTR4 (Invitrogen). This was transferred to pFGC-PPOprom to generate the final CPT-RNAi construct. The integrity of all constructs was verified by sequencing.
Agrobacterium-Mediated Transformation of T. brevicorniculatum
Agrobacterium tumefaciens strain EHA105, carrying the RNAi construct, was cultured in 100 mL of induction broth (5 g L−1 Suc, 5 g L−1 peptone, 5 g L−1 casein hydrolysate, 1 g L−1 yeast extract, 10 mm MES, and 2 mm MgSO4, pH 5.6) containing the appropriate antibiotics (50 mg L−1 kanamycin, 100 mg L−1 rifampicin, and 300 mg L−1 streptomycin). The bacteria were cultured at 28°C to the end of the log phase and then centrifuged, and the pellet was resuspended in coculture medium (4.4 g L−1 Murashige and Skoog salt solution including vitamins, 10 mm MES, and 20 g L−1 Glc, pH 5.6) supplemented with 200 μm acetosyringone. Leaf discs (approximately 1 cm2) were punched from the leaves of 6- to 10-week-old T. brevicorniculatum plants that had been grown under sterile conditions on solid medium (2.2 g L−1 Murashige and Skoog salt solution, 10 g L−1 Glc, and 8 g L−1 agar, pH 5.8) and inoculated in the coculture medium with A. tumefaciens for 30 min. The leaf discs were then placed on filter paper for 14 to 20 h at 26°C. To induce regeneration, the leaf discs were placed on regeneration medium (4.4 g L−1 Murashige and Skoog salt solution including vitamins, 18 g L−1 Glc, and 8.5 g L−1 agar, pH 5.8) supplemented with 1 mg L−1 6-benzyladenine and 0.2 mg L−1 naphthaleneacetic acid for callus and shoot induction. The elongation of shoots was maintained by the addition of 2 mg L−1 zeatin, 20 μg L−1 naphthaleneacetic acid, and 20 μg L−1 GA3. Rooting was induced on regeneration medium without hormones, and all regeneration media contained 3 mg L−1 phosphinothricin for selection. All chemicals and reagents were purchased from Duchefa. Transgenic dandelion plants were transferred to soil and cultivated at 16°C with a 16-h photoperiod.
RNA Isolation and Quantitative RT-PCR
Total RNA was extracted from latex by dissecting stems with a razor blade and harvesting 20 μL of expelling latex in 100 μL of homogenization buffer (4 m guanidine isothiocyanate, 100 mm Tris-HCl, pH 7.0, and 5 mm dithiothreitol). After adding 900 μL of QIAzol Lysis Reagent, total RNA was isolated using the RNeasy Lipid Tissue Mini Kit (Qiagen). RNA samples were treated with RNase-free DNase I (New England Biolabs), precipitated by the addition of 0.1% (v/v) 7.5 m ammonium acetate and 2.5% (v/v) 100% ethanol, and incubated at –20°C for 1 h. After centrifugation (13,000 rpm, 20 min, 4°C), RNA pellets were washed with 100 μL of 70% ethanol, dried, dissolved in 30 μL of water, and reverse transcribed with SuperScript II Reverse Transcriptase according to the manufacturer’s protocol (Invitrogen) using oligo(dT) primers. cDNA was used as the template for quantitative RT-PCR with gene-specific primers, the iCycler real-time PCR system, and the iQSYBR Green Supermix (Bio-Rad). TbCPT1 to -3 were amplified using a common forward primer, TbCPT-fwd (5′-CCACGTATCTCCTCGTATGTG-3′), in combination with the specific reverse primers TbCPT1-rev (5′-CTATTGGCCATGGCGCCACT-3′), TbCPT2-rev (5′-CTATTGGCCATGGCGCCATC-3′), and TbCTP3-rev (5′-TCCGTTCATGACCCGTATGCTT-3′). In each case, T. brevicorniculatum glycerine aldehyde-3-phosphate dehydrogenase (TbGAPDH) was used as a reference gene and was amplified using the primer combination TbGAPDH-fwd (5′-TTGGAATTGTCGAGGGTCTC-3′) and TbGAPDH-rev (5′-TGCTGCTAGGGATGATGTTG-3′). The relative expression of each target gene was calculated with REST-MCS software (Pfaffl et al., 2002).
SDS-PAGE and Western Blot
Latex was harvested from roots of adult T. brevicorniculatum plants by dissecting the tissue with a razor and transferring the expelling latex into ice-cold rubber extraction buffer (100 mm Tris, pH 7.8, 350 mm sorbitol, 10 mm NaCl, 5 mm MgCl2, and 5 mm dithiothreitol). SDS-PAGE was carried out using equal amounts of protein from whole latex protein per lane. Protein concentrations were determined using the Bradford method (Bradford, 1976). Proteins were separated on a 10% SDS polyacrylamide gel and were transferred to a nitrocellulose membrane as described previously (Towbin et al., 1979). The membrane was stained with Ponceau S to visualize the total proteins. After washing, the membrane was incubated with the primary α-CPT antibody (1:250 dilution) for 1 h at room temperature, washed, and then incubated with the secondary α-rabbit IgG antibody (1:10,000 dilution) conjugated to horseradish peroxidase (Sigma) according to the manufacturer’s instructions. The primary antibody recognizes all three TbCPTs and was generated as described previously (Schmidt et al., 2010b). The horseradish peroxidase-coupled secondary antibody signal was imaged on x-ray films by chemiluminescence detection using SuperSignal West Dura Extended Duration Substrate (Thermo Scientific). Signal intensities were quantified using the ImageJ software (http://rsbweb.nih.gov) and normalized to the Ponceaus S signal of the corresponding samples.
[1-14C]IPP Incorporation Assay
To measure the [14C]IPP incorporation rate, crude latex was harvested and transferred into ice-cold rubber extraction buffer, and 50 μg of whole latex protein was used in each assay as described by Schmidt et al. (2010b). The reaction contained 2.8 μm E,E-FPP (Sigma) and 7.2 μm (16.7 kBq) [1-14C]IPP (GE Healthcare) as substrates in a total volume of 200 μL and was incubated at 30°C for 2 h, then stopped by heating to 95°C for 5 min. The negative control contained only E,E-FPP and [14C]IPP as substrates. A two-step differential solvent extraction of radiolabeled products was performed according to Asawatreratanakul et al. (2003) with slight modifications. Short- and medium-chain polyprenyl diphosphate products were extracted twice with 200 μL of n-butanol, then long-chain products were extracted twice with 200 μL of toluene:hexane (1:1, v/v). Radioactivity was measured using a scintillation counter (Beckman Scintillation Counter LS6500) after mixing 50 μL of the extracts with 4 mL of Rotiszint eco plus (Roth). The total radioactivity of the respective complete solvent fractions was calculated taking the dilution into account.
To determine the molecular size distribution of the incorporation assay products by SEC, the assay reaction was performed as described above and the incorporation assay products were hydrolyzed with potato (Solanum tuberosum) acid phosphatase after heating to 95°C for 5 min. The corresponding alcohols were extracted with n-hexane, air dried, and redissolved in 500 μL of toluene. After filtration through a membrane with 0.2-μm pore size, the samples were air dried, redissolved in 100 μL of toluene, and 90 μL was then fractionated by SEC. Radioactivity in the different fractions was measured using a scintillation counter after mixing the whole fraction (1 mL) with 4 mL of Rotiszint eco plus.
Separation of Rubber Particles
To visualize rubber particles, 50 μL of latex was harvested from the roots of adult T. brevicorniculatum plants and transferred into 100 μL of ice-cold rubber extraction buffer. After centrifugation (13,000 rpm, 20 min, 4°C), the latex separated into three fractions: the pellet, C-serum, and an upper rubber phase containing the rubber particles.
Determination of Dry Rubber Content by Gravimetric Analysis
Whole roots were freeze dried and pulverized, and 1.5 g of root material was mixed with 80 mL of toluene, incubated at 85°C for 20 h, and centrifuged at 2,000 rpm for 10 min at room temperature. The toluene supernatant was transferred to a fresh tube, the toluene was vaporized, and the remaining sample was redissolved in 5 mL of toluene. Long-chain poly(cis-1,4-isoprene) was precipitated by incubating with 2 volumes of methanol for 30 min. After centrifugation (4,000 rpm, 60 min at room temperature), the precipitated polyisoprene appeared as a floating layer. This was washed with acetone and water for 40 min to remove resins and dried for 16 h at room temperature before weighing.
Measurement of Molecular Mass Distribution by SEC
Latex was harvested from roots of adult T. brevicorniculatum plants and transferred to 1 mL of ice-cold rubber extraction buffer in a 15-mL glass jar. Each sample was lyophilized, dissolved in 2 mL of methanol, and heated at 60°C in a water bath for 30 min. After centrifugation (2,000 rpm, 10 min), the methanol supernatant was discarded and the remaining sample was extracted twice with 2 mL of hexane at room temperature. The hexane phases were pooled and evaporated to dryness. The samples were then dissolved in toluene to a final concentration of 1 mg mL−1. Molecular mass distribution was measured using the SECcurity GPC system (Polymer Standards Service) fitted with a PSS SDV 20μ 8- × 50-mm precolumn, a PSS SDV Linear XL 20μ 8- × 300-nm analytical column, and a PSS SDV 100 Å 2 0μ 8- × 300-mm analytical column. Toluene was used as the solvent, and the system was calibrated with poly(cis-1,4-isoprene) standards from 4,470 to 999,000 D (Polymer Standards Service).
TEM Analysis
Root segments were fixed in 2.5% glutaraldehyde in phosphate-buffered saline (PBS; pH 7.3) for 45 min. After three 15-min washes in PBS, the tissue was postfixed with 1% osmium tetroxide in PBS (pH 7.3). Water was removed by immersion in a graded water:ethanol series (30%, 50%, 70%, 90%, and 96% ethanol and absolute ethanol), each step lasting 15 min. The root material was then embedded in Spurr resin by a graded Spurr:propylene oxide series (1:3, 1:1, 3:1), each step lasting 4 h. Samples were incubated in Spurr without propylene oxide for 4 h, Spurr was replaced, and samples were incubated for 12 h. Embedding was carried out with fresh Spurr that was polymerized at 70°C for 8 h. Thin sections (approximately 70 nm) were prepared on an Ultracut apparatus (Leica) using a diamond knife, placed on a copper mesh, and stained with 2.5% uranyl acetate. TEM was carried out using a Philips CM 10 (acceleration voltage of 80 kV) in bright-field mode.
Quantitative Determination of Poly(cis-1,4-Isoprene) Levels by 1H-NMR Spectroscopy
We incubated 200 mg of powdered and freeze-dried roots from 3-month-old T. brevicorniculatum plants in 5 mL of hexane for 24 h at 20°C with gentle agitation. Cell debris was removed by centrifugation (3,000 rpm, 5 min) and filtration. Hexane was removed under reduced pressure. The residue was dissolved in 550 μL of deuterated dichloromethane (CD2Cl2; same batch for each sample) and analyzed by 1H-NMR spectroscopy using a digital Bruker AVANCE-I 500 MHz spectrometer equipped with an inverse 5-mm H/C probe head, temperature control (battery voltage temperature; BCU-05 unit), and a sample changer. All measurements were obtained in full automation mode using ICONNMR and TOPSPIN 2.3 (Bruker). The temperature was equilibrated to 300 K inside the magnet for 5 min before measurement commenced. For each sample, the probe head was tuned and matched using the ATMA function. Each sample was shimmed with the TOPSHIM program. The 1H-NMR data were then acquired using a one-dimensional 1H-NMR pulse program with a 30°C pulse and a relaxation delay of 5 s.
Prior to Fourier transformation, the FID was zero filled to 128 k and multiplied with an exponential window function (line-broadening function, 0.2). The phase and the baseline were corrected carefully. The residual CHDCl2 signals of the deuterated solvent (triplet, 5.32 ppm) and the C5 methyl signal for poly(cis-1,4-isoprene) at 1.66 ppm (well-resolved singlet) were manually integrated using the same limits for each spectrum. The integral value of the solvent was set to 1.0. To assess the amount of poly(cis-1,4-isoprene) in each sample, a reference sample containing 2 mg of poly(cis-1,4-isoprene) in 550 μL of CD2Cl2 (same batch as before) was prepared, measured, and analyzed as described above. The 1H integral of the C5 poly(cis-1,4-isoprene) signal at 1.66 ppm (relative to an integral value of 1.0 for the solvent signal) was then used to normalize the concentration of poly(cis-1,4-isoprene) in the other extracts.
Triterpene Analysis
Whole roots from 3-month-old plants were frozen in liquid nitrogen, freeze dried, and crushed to a fine powder. We used 100 mg of dry root material for saponification by adding 3 mL of methanol containing 6% potassium hydroxide and heating to 75°C for 2 h. Evaporated methanol was refilled when necessary. The samples were then cooled to room temperature and mixed with 1 volume of water. Samples were extracted three times with 1 volume of hexane for 60 min at room temperature. Hexane phases were pooled, spiked with betuline as an internal standard, evaporated to dryness, and the residue was acetylated in 100 μL of toluene containing acetic anhydride and pyridine for 1 h at 70°C. The reagents were evaporated under an air stream and then completely desiccated by freeze drying. Pentacyclic triterpenes and sterols were quantified with a 3400 Varian GC/FID equipped with a DB5 column (J&W; 30 m long, 0.32 mm i.d., 0.25 μm film thickness). Compounds were identified by gas chromatography-mass spectrometry (GC-MS) using a 6890 Agilent gas chromatograph equipped with a DB5-MS column coupled to a 5973 mass selective detector. The GC programs included a steep ramp from 60°C to 220°C at 30°C min−1 and then from 220°C to 300°C at 2°C min−1. Pentacyclic triterpenes were identified based on previous studies (Shiojima et al., 1992; Kushiro et al., 2000; Husselstein-Muller et al., 2001).
HMGR Activity Assay
Crude latex was harvested and transferred into ice-cold rubber extraction buffer. Approximately 200 μg of whole protein was used in the [3-14C]HMG-CoA incorporation assay as described by Bach et al. (1986). After lactonization, the samples were centrifuged (13,000 rpm, 1 min), and 15 μL of the supernatant was analyzed by silica gel thin-layer chromatography (silica gel 60 TLC plates; Merck) using 1:1 toluene:acetone as the solvent. Bands representing labeled mevalonolactone were localized by phosphor imaging (Typhoon; Amersham Bioscience) and cut from the sheet so that the incorporation of radioactivity into mevalonolactone could be measured by liquid scintillation counting using Rotiszint eco plus (Roth). The HMGR activity was determined as the amount of labeled HMG-CoA incorporated into mevalonolactone mg−1 protein h−1.
Quantitative Determination of Inulin Levels by 1H-NMR Spectroscopy
We mixed 12 mg of powdered and freeze-dried roots from 3-month-old T. brevicorniculatum plants with 0.8 g of glass beads (0.25–0.50 mm diameter; Roth) and 1 mL of deuterated water at 70°C. The mixture was immediately homogenized (Ribolyser; three 20-s cycles at 6.5 m s−1) and incubated at 70°C for 10 min to allow the quantitative dissolution of inulin in the hot solvent. Cell debris and the glass beads were separated by centrifugation (5,000 rpm, 10 min), and 550 μL of the supernatant was analyzed by 1H-NMR spectroscopy. We added 10 μL of deuterated [2,2,3,3-2H4]3-(trimethylsilyl)propionic acid, sodium salt (TSP; 6.037 mm in D2O) as an internal standard. The final concentration of TSP was 0.108 mm (corresponding to 10.4 μg) in each of the samples. The solutions were immediately analyzed by 1H-NMR spectroscopy in triplicate as described above, and 1H-NMR data were acquired with presaturation of the residual water signal using a one-dimensional NOESY pulse program with gradient selection and phase compensation.
Prior to Fourier transformation, the FID was zero filled to 128 k and multiplied with an exponential window function (line-broadening function, 0.2). The phase and baseline were corrected carefully. Signals representing TSP (singlet, 0 ppm) and inulin (well-resolved triplet of H4 at 4.04 ppm) were manually integrated using the same limits for each spectrum. The integral value of TSP was set to 1.0. The sd values between the integrals of the technical replicates were less than 2%, demonstrating the robustness of the method. Averages from three extracts generated sd values of less than 20%. To determine the absolute amounts of inulin, we prepared three reference samples containing 8 mg of inulin and 0.108 mm deuterated TSP in 560 μL of D2O, measured and analyzed using the protocols described above. The averaged relative integral values (relative to 1.0 for the TSP signal) of the inulin triplet at 4.04 ppm were then used to reference the inulin concentration in the other extracts.
Quantitative Determination of Inulin Levels by Hydrolysis
We obtained root extracts by milling with glass beads (see above), mixed 100 μL of each extract with 100 μL of 50 mm HCl in triplicate, and incubated the reactions for 45 min at 100°C. Free Fru and Glc levels were then measured in an assay mixture comprising 20 μL of each hydrolysate, 300 mm triethanolamine buffer (pH 7.6), 35 mm MgSO4, 0.5 mm NADP, 1.7 mm ATP, 2.2 units of Glc-6-P dehydrogenase, 2.2 units of hexokinase, and 5 units of Glc-6-P isomerase (total volume, 1.4 mL). The mixture was incubated at 20°C for 10 min, and the NADPH generated in the reaction was measured by spectrophotometry at 366 nm with two technical replicates using a molar extinction coefficient of 3.5 L mmol−1 cm−1. The resulting values were corrected for the amount of free Fru, Glc, and Suc by concentrating 100 μL of root extract (see above) and 10 μL of 10 mm ribitol (internal standard) to dryness under a stream of nitrogen gas (three technical replicates for each sample), dissolving the residue in 100 μL of pyridine containing 40 mg mL−1 methoxyamine hydrochloride and incubating for 90 min at 35°C. The reaction mixture was dried under nitrogen, and the residue was dissolved in 100 μL of N-methyl-N-trimethylsilyltrifluoracetamide containing 1% trimethylchlorsilane and heated for 30 min at 50°C. The mixtures were analyzed by GC-MS using a QP2010 Plus (Shimadzu) on a 30 m Equity-5 column (Supelco) in split mode 1:20, using a 150°C to 290°C temperature gradient at 5°C min−1, and the detector and injector temperature set to 260°C. Each analysis was performed with two biological replicates.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. RNA silencing of TbCPT1 to -3 in latex from petioles.
Supplemental Figure S2. Structures of poly(cis-1,4-isoprene) (n > 10,000), triterpene compounds, and inulin (n = 10–100) in T. brevicorniculatum.
Supplemental Table S1. Concentrations of Fru, Glc, and Suc in the roots of T. brevicorniculatum plants as determined by spectrophotometry and GC-MS before and after acidic hydrolysis.
Acknowledgments
We gratefully acknowledge the technical assistance of Sandra Ponanta, Daniela Ahlert, and Ursula Malkus (Westphalian Wilhelms-University of Münster) and Christine Schwarz (Technische Universität München).
References
- Akashi T, Furuno T, Takahashi T, Ayabe SI. (1994) Biosynthesis of triterpenoids in cultured cells, and regenerated and wild plant organs of Taraxacum officinale. Phytochemistry 36: 303–308 [Google Scholar]
- Asawatreratanakul K, Zhang YW, Wititsuwannakul D, Wititsuwannakul R, Takahashi S, Rattanapittayaporn A, Koyama T. (2003) Molecular cloning, expression and characterization of cDNA encoding cis-prenyltransferases from Hevea brasiliensis: a key factor participating in natural rubber biosynthesis. Eur J Biochem 270: 4671–4680 [DOI] [PubMed] [Google Scholar]
- Bach TJ, Rogers DH, Rudney H. (1986) Detergent-solubilization, purification, and characterization of membrane-bound 3-hydroxy-3-methylglutaryl-coenzyme A reductase from radish seedlings. Eur J Biochem 154: 103–111 [DOI] [PubMed] [Google Scholar]
- Bandurski RS, Teas HJ. (1957) Rubber biosynthesis in latex of Hevea brasiliensis. Plant Physiol 32: 643–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner J. (1991) The history of rubber. In JW Whitworth, EE Whitehead, eds, Guayule Natural Rubber: A Technical Publication with Emphasis on Recent Findings. Guayule Administrative Management Committee and US Department of Agriculture Cooperative State Research Service, Office of Arid Lands Studies, University of Arizona, Tucson, AZ, pp 1–16 [Google Scholar]
- Bouvier F, Rahier A, Camara B. (2005) Biogenesis, molecular regulation and function of plant isoprenoids. Prog Lipid Res 44: 357–429 [DOI] [PubMed] [Google Scholar]
- Bouvier-Navé P, Berna A, Noiriel A, Compagnon V, Carlsson AS, Banas A, Stymne S, Schaller H. (2010) Involvement of the phospholipid sterol acyltransferase1 in plant sterol homeostasis and leaf senescence. Plant Physiol 152: 107–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Bushman BS, Scholte AA, Cornish K, Scott DJ, Brichta JL, Vederas JC, Ochoa O, Michelmore RW, Shintani DK, Knapp SJ. (2006) Identification and comparison of natural rubber from two Lactuca species. Phytochemistry 67: 2590–2596 [DOI] [PubMed] [Google Scholar]
- Cérantola S, Kervarec N, Pichon R, Magné C, Bessieres MA, Deslandes E. (2004) NMR characterisation of inulin-type fructooligosaccharides as the major water-soluble carbohydrates from Matricaria maritima (L.). Carbohydr Res 339: 2445–2449 [DOI] [PubMed] [Google Scholar]
- Chappell J, Wolf F, Proulx J, Cuellar R, Saunders C. (1995) Is the reaction catalyzed by 3-hydroxy-3-methylglutaryl coenzyme A reductase a rate-limiting step for isoprenoid biosynthesis in plants? Plant Physiol 109: 1337–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Closa M, Vranová E, Bortolotti C, Bigler L, Arró M, Ferrer A, Gruissem W. (2010) The Arabidopsis thaliana FPP synthase isozymes have overlapping and specific functions in isoprenoid biosynthesis, and complete loss of FPP synthase activity causes early developmental arrest. Plant J 63: 512–525 [DOI] [PubMed] [Google Scholar]
- Cornish K. (2001) Similarities and differences in rubber biochemistry among plant species. Phytochemistry 57: 1123–1134 [DOI] [PubMed] [Google Scholar]
- Cornish K, Wood DF, Windle JJ. (1999) Rubber particles from four different species, examined by transmission electron microscopy and electron-paramagnetic-resonance spin labeling, are found to consist of a homogeneous rubber core enclosed by a contiguous, monolayer biomembrane. Planta 210: 85–96 [DOI] [PubMed] [Google Scholar]
- Cunillera N, Arró M, Delourme D, Karst F, Boronat A, Ferrer A. (1996) Arabidopsis thaliana contains two differentially expressed farnesyl-diphosphate synthase genes. J Biol Chem 271: 7774–7780 [DOI] [PubMed] [Google Scholar]
- Cunillera N, Arró M, Forés O, Manzano D, Ferrer A. (2000) Characterization of dehydrodolichyl diphosphate synthase of Arabidopsis thaliana, a key enzyme in dolichol biosynthesis. FEBS Lett 477: 170–174 [DOI] [PubMed] [Google Scholar]
- Cunillera N, Boronat A, Ferrer A. (1997) The Arabidopsis thaliana FPS1 gene generates a novel mRNA that encodes a mitochondrial farnesyl-diphosphate synthase isoform. J Biol Chem 272: 15381–15388 [DOI] [PubMed] [Google Scholar]
- Furuno T, Kamiyama A, Akashi T, Usui M, Takahashi T, Ayabe S-I. (1993) Triterpenoid constituents of tissue cultures and regenerated organs of Taraxacum officinale. Plant Tissue Cult Lett 10: 275–280 [Google Scholar]
- Hepper CM, Audley BG. (1969) The biosynthesis of rubber from beta-hydroxy-beta-methylgluarylcoenzyme A in Hevea brasiliensis latex. Biochem J 114: 379–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AHC. (1992) Oil bodies and oleosins in seeds. Annu Rev Plant Physiol Plant Mol Biol 43: 177–200 [Google Scholar]
- Husselstein-Muller T, Schaller H, Benveniste P. (2001) Molecular cloning and expression in yeast of 2,3-oxidosqualene-triterpenoid cyclases from Arabidopsis thaliana. Plant Mol Biol 45: 75–92 [DOI] [PubMed] [Google Scholar]
- Kang H, Kang MY, Han KH. (2000) Identification of natural rubber and characterization of rubber biosynthetic activity in fig tree. Plant Physiol 123: 1133–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharel Y, Koyama T. (2003) Molecular analysis of cis-prenyl chain elongating enzymes. Nat Prod Rep 20: 111–118 [DOI] [PubMed] [Google Scholar]
- Kushiro T, Shibuya M, Masuda K, Ebizuka Y. (2000) A novel multifunctional triterpene synthase from Arabidopsis thaliana. Tetrahedron Lett 41: 7705–7710 [Google Scholar]
- Lange BM, Ghassemian M. (2003) Genome organization in Arabidopsis thaliana: a survey for genes involved in isoprenoid and chlorophyll metabolism. Plant Mol Biol 51: 925–948 [DOI] [PubMed] [Google Scholar]
- Langenheim JH. (2003) Plant Resins: Chemistry, Evolution, Ecology and Ethnobotany. Timber Press, Portland, OR, pp 1223–1280 [Google Scholar]
- Lu YP, Liu HG, Teng KH, Liang PH. (2010) Mechanism of cis-prenyltransferase reaction probed by substrate analogues. Biochem Biophys Res Commun 400: 758–762 [DOI] [PubMed] [Google Scholar]
- Manzano D, Busquets A, Closa M, Hoyerová K, Schaller H, Kamínek M, Arró M, Ferrer A. (2006) Overexpression of farnesyl diphosphate synthase in Arabidopsis mitochondria triggers light-dependent lesion formation and alters cytokinin homeostasis. Plant Mol Biol 61: 195–213 [DOI] [PubMed] [Google Scholar]
- Martin S, Parton RG. (2006) Lipid droplets: a unified view of a dynamic organelle. Nat Rev Mol Cell Biol 7: 373–378 [DOI] [PubMed] [Google Scholar]
- Mooibroek H, Cornish K. (2000) Alternative sources of natural rubber. Appl Microbiol Biotechnol 53: 355–365 [DOI] [PubMed] [Google Scholar]
- Munt O, Arias M, Hernandez M, Ritter E, Schulze Gronover C, Prüfer D. (2012) Fertilizer and planting strategies to increase biomass and improve root morphology in the natural rubber producer Taraxacum brevicorniculatum. Ind Crops Prod 36: 289–293 [Google Scholar]
- Nasab EE, Habibi-Rezaei M, Khaki A, Balvardi M. (2009) Investigation on acid hydrolysis of inulin: a response surface methodology approach. Int J Food Eng 5: Article 12 [Google Scholar]
- Nguyen SK, Sophonputtanaphoca S, Kim E, Penner MH. (2009) Hydrolytic methods for the quantification of fructose equivalents in herbaceous biomass. Appl Biochem Biotechnol 158: 352–361 [DOI] [PubMed] [Google Scholar]
- Oh SK, Han KH, Ryu SB, Kang H. (2000) Molecular cloning, expression, and functional analysis of a cis-prenyltransferase from Arabidopsis thaliana: implications in rubber biosynthesis. J Biol Chem 275: 18482–18488 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punetha A, Muthukumaran J, Hemrom AJ, Arumugam N. (2010) Towards understanding the regulation of rubber biosynthesis: insights into the initiator and elongator enzymes. J Bioinform Seq Anal 2: 001–010 [Google Scholar]
- Sando T, Takeno S, Watanabe N, Okumoto H, Kuzuyama T, Yamashita A, Hattori M, Ogasawara N, Fukusaki E, Kobayashi A. (2008) Cloning and characterization of the 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway genes of a natural-rubber producing plant, Hevea brasiliensis. Biosci Biotechnol Biochem 72: 2903–2917 [DOI] [PubMed] [Google Scholar]
- Sato M, Fujisaki S, Sato K, Nishimura Y, Nakano A. (2001) Yeast Saccharomyces cerevisiae has two cis-prenyltransferases with different properties and localizations: implication for their distinct physiological roles in dolichol synthesis. Genes Cells 6: 495–506 [DOI] [PubMed] [Google Scholar]
- Schaller H, Grausem B, Benveniste P, Chye M-L, Tan C-T, Song Y-H, Chua N-H. (1995) Expression of the Hevea brasiliensis (H.B.K.) Müll. Arg. 3-hydroxy-3-methylglutaryl-coenzyme A reductase 1 in tobacco results in sterol overproduction. Plant Physiol 109: 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T, Hillebrand A, Wurbs D, Wahler D, Lenders M, Schulze Gronover C, Prüfer D. (2010a) Molecular cloning and characterisation of rubber biosynthetic genes from Taraxacum koksaghyz. Plant Mol Biol Rep 28: 277–284 [Google Scholar]
- Schmidt T, Lenders M, Hillebrand A, van Deenen N, Munt O, Reichelt R, Eisenreich W, Fischer R, Prüfer D, Gronover CS. (2010b) Characterization of rubber particles and rubber chain elongation in Taraxacum koksaghyz. BMC Biochem 11: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütz K, Muks E, Carle R, Schieber A. (2006) Separation and quantification of inulin in selected artichoke (Cynara scolymus L.) cultivars and dandelion (Taraxacum officinale WEB. ex WIGG.) roots by high-performance anion exchange chromatography with pulsed amperometric detection. Biomed Chromatogr 20: 1295–1303 [DOI] [PubMed] [Google Scholar]
- Seetang-Nun Y, Sharkey TD, Suvachittanont W. (2008) Molecular cloning and characterization of two cDNAs encoding 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Hevea brasiliensis. J Plant Physiol 165: 991–1002 [DOI] [PubMed] [Google Scholar]
- Shiojima K, Arai Y, Masuda K, Takase Y, Ageta T, Ageta H. (1992) Mass spectra of pentacyclic triterpenoids. Chem Pharm Bull (Tokyo) 40: 1683–1690 [Google Scholar]
- Siler DJ, Cornish K. (1993) A protein from Ficus elastica rubber particles is related to proteins from Hevea brasiliensis and Parthenium argentatum. Phytochemistry 32: 1097–1102 [Google Scholar]
- Stermer BA, Bianchini GM, Korth KL. (1994) Regulation of HMG-CoA reductase activity in plants. J Lipid Res 35: 1133–1140 [PubMed] [Google Scholar]
- Surmacz L, Swiezewska E. (2011) Polyisoprenoids: secondary metabolites or physiologically important superlipids? Biochem Biophys Res Commun 407: 627–632 [DOI] [PubMed] [Google Scholar]
- Szambelan K, Nowak J. (2006) Acidic and enzymatic hydrolysis of Jerusalem artichoke (Helianthus tuberosus L.) tubers for further ethanol production. Electron J Pol Agric Univ 9: Article 38 [Google Scholar]
- Takahashi S, Koyama T. (2006) Structure and function of cis-prenyl chain elongating enzymes. Chem Rec 6: 194–205 [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 76: 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beilen JB, Poirier Y. (2007) Guayule and Russian dandelion as alternative sources of natural rubber. Crit Rev Biotechnol 27: 217–231 [DOI] [PubMed] [Google Scholar]
- van Deenen N, Bachmann A-L, Schmidt T, Schaller H, Sand J, Prüfer D, Schulze Gronover C. (August 11, 2011) Molecular cloning of mevalonate pathway genes from Taraxacum brevicorniculatum and functional characterisation of the key enzyme 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Mol Biol Rep http://dx.doi.org/10.1007/s11033-011-1221-4 [DOI] [PubMed] [Google Scholar]
- Van den Ende W, Michiels A, Van Wonterghem D, Vergauwen R, Van Laere A. (2000) Cloning, developmental, and tissue-specific expression of sucrose:sucrose 1-fructosyl transferase from Taraxacum officinale: fructan localization in roots. Plant Physiol 123: 71–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahler D, Gronover CS, Richter C, Foucu F, Twyman RM, Moerschbacher BM, Fischer R, Muth J, Prüfer D. (2009) Polyphenoloxidase silencing affects latex coagulation in Taraxacum species. Plant Physiol 151: 334–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentzinger LF, Bach TJ, Hartmann MA. (2002) Inhibition of squalene synthase and squalene epoxidase in tobacco cells triggers an up-regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase. Plant Physiol 130: 334–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood DF, Cornish K. (2000) Microstructure of purified rubber particles. Int J Plant Sci 161: 435–445 [DOI] [PubMed] [Google Scholar]
- Zhang H, Ohyama K, Boudet J, Chen Z, Yang J, Zhang M, Muranaka T, Maurel C, Zhu J-K, Gong Z. (2008) Dolichol biosynthesis and its effects on the unfolded protein response and abiotic stress resistance in Arabidopsis. Plant Cell 20: 1879–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]