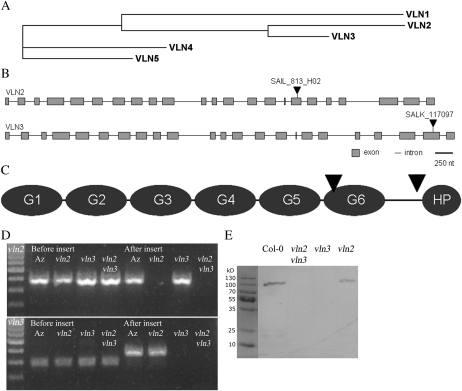
Figure 1.
Characterization of the Arabidopsis villin gene family and T-DNA insertions in vln2 and vln3. A, Cladogram of the Arabidopsis villins, based on cDNA sequences. B, Locations of T-DNA inserts in vln2 and vln3. Gray boxes represent exons, and horizontal lines represent introns. T-DNA inserts (arrowheads) are not drawn to scale. C, Domain structure of villin. Arrowheads show locations corresponding to the locations of T-DNA inserts of vln2 and vln3. D, T-DNA insertions result in truncated transcripts in vln2, vln3, and vln2 vln3. Products could be amplified using a cDNA template using VLN-specific primers before the inserts, but when both primers (vln2) or the reverse primer (vln3) were designated for coding regions after the insert (Supplemental Fig. S3; Supplemental Table S1), products could not be amplified. Az, Azygous. E, A protein gel blot of Col-0, vln2, vln3, and vln2 vln3 root extracts probed with lily anti-villin antibody (Tominaga et al., 2000) shows that vln3 and vln2 vln3 do not contain (a truncated version of) the VLN3 protein.