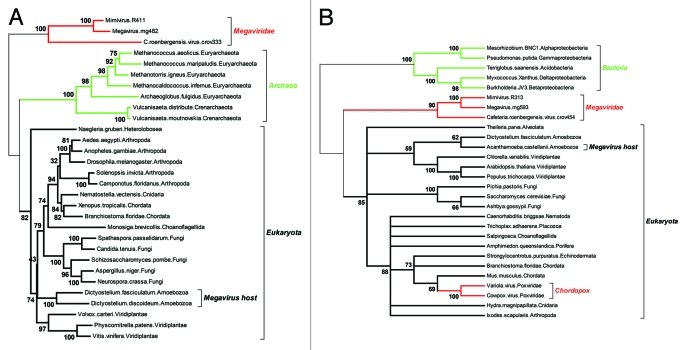
Figure 2.
Two reliable phylogenetic reconstructions positionning the Megavirus in a partial Tree of Life. As the quality of the multiple alignment is essential to the reliability of the derived phylogeny, we only included the most similar proteins sequences of each clade in the analyses. (A) Positionning of the three closest Megavirus relatives using the largest clamp loader subunits. The multiple alignment (default options) and tree reconstruction (neighbor joining on 312 ungapped position, JTT substitution model) was performed using the on the MAFFT server (mafft.cbrc.jp/alignment/server/). The highly divergent bacterial homologs were not included, to preserve the quality of the multiple alignment. The deepest bootstrap values indicate the total lack of affinity of the Megaviridae sequences with both the archaeal and eukaryotic domains. (B) Positionning of the three closest Megavirus relatives using their largest ribonucleoside diphosphate reductase subunits. The multiple alignment (default options) and tree reconstruction (neighbor joining on 735 ungapped position, JTT substitution model) was performed as above. This time, the highly divergent archaeal homologs were not included, to preserve the quality of the multiple alignment. The deepest bootstrap values indicate a total lack of affinity of the Megaviridae with the bacterial and eukaryotic domains. In contrast, the acquisition of the vertebrate gene by the chordopoxviruses is showing very clearly, serving as an internal control that virus genes acquired by lateral transfer are indeed detectable. Amoebozoa sequences are indicated in A) and B) to emphasize that the Megavirus/Mimivirus genes do not cluster with their host’s homologs.