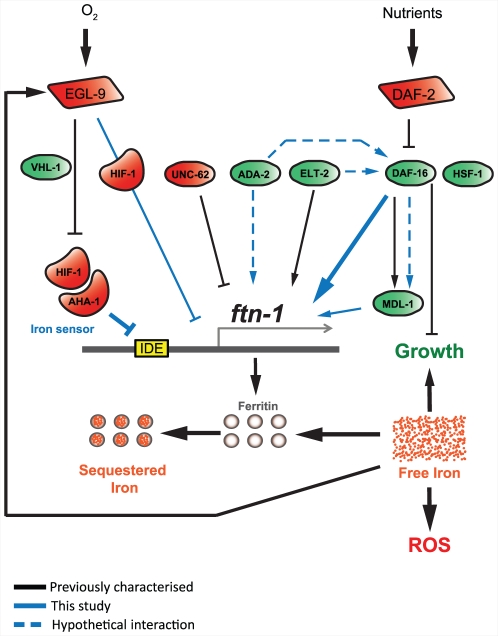
Figure 7. ftn-1 expression is regulated by both the insulin/IGF-1 and hypoxia signaling pathways.
This figure provides a diagrammatic representation of the gene regulatory networking controlling ftn-1 expression. It includes previously established regulatory elements (black lines), newly established regulatory elements (blue lines) and new, hypothetical regulatory elements (dashed blue lines). We identified a positive regulatory role for the genes mdl-1, hsf-1, ada-2 and daf-16. In the case of mdl-1, previous work suggests that this transcription factor acts downstream of DAF-16, but it is unclear whether this is true for ftn-1 regulation (hence second line, dotted blue, from DAF-16 to MDL-1), or whether MDL-1 acts independently of DAF-16 in this case. Loss of ada-2 or elt-2 reduces ftn-1 expression but we were unable to detect an effect of ada-2 or elt-2 RNAi in the absence of DAF-16. While this may be caused by a lack of sensitivity in our assay, it could also indicate that these factors may act together with DAF-16 or upstream of DAF-16 to regulate ftn-1 expression. We found that hif-1 and aha-1 repress ftn-1 expression and that hif-1 is required for iron-dependent regulation of ftn-1, implying that HIF acts as an iron sensor in C. elegans. However, HIF-1 activity on ftn-1 expression can be regulated through both vhl-1-dependent and independent pathways and our data shows that these pathways act antagonistically on ftn-1 expression. The VHL-1-independent inhibition of ftn-1 expression by EGL-9 could either involve activation of transcriptional repression by HIF-1 or (more parsimoniously) inhibition of transcriptional activation by HIF-1. The latter interpretation would suggest the presence of a co-regulator that turns HIF-1 into a transcriptional activator of ftn-1.