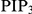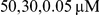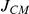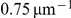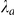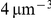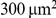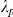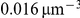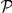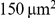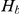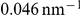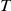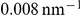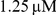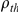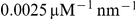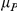
|
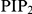 -dependent Arp2/3 activation rate -dependent Arp2/3 activation rate |
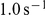
|
[25]
|
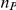
|
Hill coeff. of 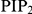 -mediated Arp2/3 activation -mediated Arp2/3 activation |

|
[25]
|
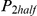
|
Threshold conc. of 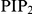 for Arp2/3 activation for Arp2/3 activation |
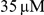
|
[25]
|
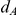
|
Arp2/3 decay rate |
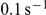
|
[36]
|
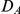
|
Arp2/3 diffusion coefficient |
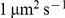
|
[93]
|
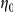
|
Arp2/3 nucleation rate |
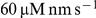
|
[36]
|
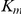
|
saturation constant for Arp2/3 nucleation |
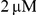
|
[36]
|
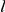
|
scale factor (converts units of 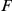 to conc.) to conc.) |
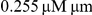
|
[36]
|
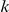
|
scale factor (converts conc. to units of 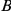 ) ) |
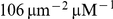
|
[36]
|
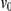
|
actin filament growth rate (free polymerization) |
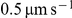
|
[94], [95]
|
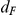
|
actin filament turnover rate |
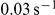
|
[88], [96]
|
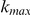
|
barbed end capping rate |
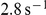
|
[95], [97]
|
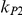
|
max reduction of capping by 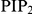
|
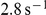
|
[25]
|

|
reduction in capping rate near leading edge |
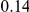
|
[36]
|
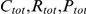
|
total levels of Cdc42, Rac, Rho |
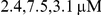
|
[36], [91], [98]
|
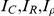
|
Cdc42, Rac, Rho activation input rates |
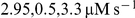
|
[36], [91]
|
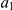
|
Rho level for half-max inhibition of Cdc42 |
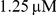
|
[36]
|
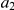
|
Cdc42 level for half-max inhibition of Rho |
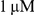
|
[36]
|

|
Hill coeff. of Cdc42-Rho mutual inhibition |

|
[36]
|

|
Cdc42-dependent Rac activation rate |
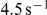
|
[36]
|
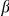
|
Rac-dependent Rho activation rate |
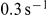
|
[36]
|
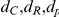
|
decay rates of activated Rho-proteins |
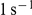
|
[99], [100]
|
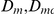
|
diffusion coeff. of active, inactive Rho-proteins |
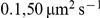
|
[62], [101]
|
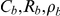
|
typical basal levels of active Cdc42, Rac, Rho |
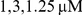
|
[36]
|
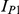
|
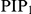 input rate input rate |
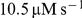
|
[92]
|
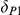
|
PIP decay rate decay rate |
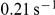
|
[92]
|
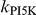
|
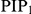 to to 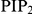 baseline conversion rate (by PI5K) baseline conversion rate (by PI5K) |
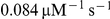
|
[92]
|
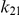
|
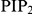 to to 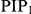 conversion rate conversion rate |
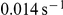
|
[92]
|
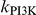
|
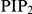 to to 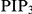 baseline conversion rate (by PI3K) baseline conversion rate (by PI3K) |
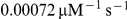
|
[92]
|
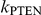
|
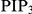 to to 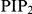 baseline conversion rate (by PTEN) baseline conversion rate (by PTEN) |
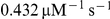
|
[92]
|
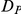
|
PI diffusion rate |
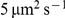
|
[101], [102]
|
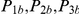
|
typical basal levels of 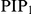 , , 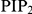 , , 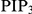
|
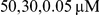
|
[70], [103]
|
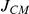
|
coupling energy per boundary site |
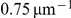
|
[36]
|
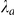
|
cell inelasticity |
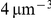
|
[36]
|
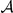
|
target area of the cell |
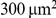
|
[36]
|
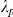
|
membrane inelasticity |
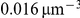
|
–– |
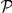
|
target perimeter of the cell |
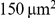
|
–– |
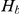
|
membrane yield |
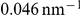
|
[36]
|
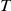
|
simulation “temperature” |
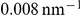
|
[36]
|

|
effect of Rho on contraction |
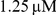
|
[36]
|
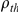
|
Rho contraction threshold |
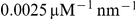
|
[36]
|
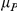
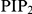 -dependent Arp2/3 activation rate
-dependent Arp2/3 activation rate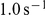
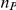
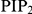 -mediated Arp2/3 activation
-mediated Arp2/3 activation
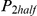
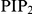 for Arp2/3 activation
for Arp2/3 activation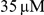
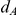
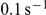
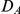
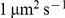
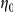
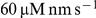
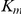
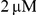
 to conc.)
to conc.)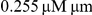
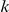
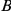 )
)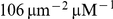
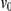
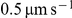
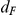
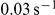
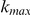
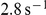
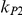
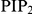
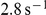

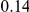
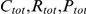
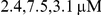
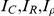
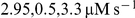
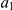
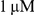



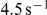
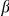
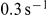
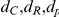
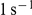
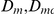
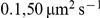
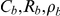
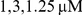
 input rate
input rate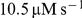
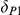
 decay rate
decay rate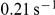
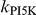
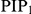 to
to 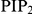 baseline conversion rate (by PI5K)
baseline conversion rate (by PI5K)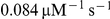
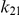
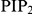 to
to 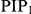 conversion rate
conversion rate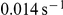
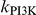
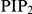 to
to 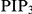 baseline conversion rate (by PI3K)
baseline conversion rate (by PI3K)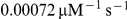
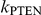
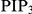 to
to 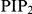 baseline conversion rate (by PTEN)
baseline conversion rate (by PTEN)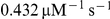
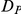
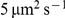
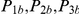
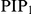 ,
, 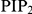 ,
,